You are here
Covid-19 variants are a game-changer
Although vaccination campaigns were meant to accelerate the end of the epidemic, the appearance of different variantsFermerAn organism that differs from the original virus because of one or more mutations. When these mutations modify its impact (in terms of transmissibility, virulence or immune escape), it is referred to as a “variant of interest." of the Covid-19 virus is raising fears that it might resume with even greater intensity. Although this is bad timing, the emergence of mutations remains a natural phenomenon that is increasingly likely to occur as an epidemic spreads. What then makes the UK, South African and Brazilian variants distinct from the original strains?
“There is often confusion about the terminology, which can lead to incorrect information,” points out Bruno Lina, university professor and scientist at the International Centre for Infectiology Research (CIRI),1 practitioner at the Hospices Civils de Lyon university hospital, director of France’s national reference centre (CNR) for infectious respiratory viruses and a member of the independent scientific board in charge of advising the French government on the epidemic. “Covid-19 is caused by a single virus: SARS-CoV-2. It evolves according to different lineages which, after changes to their genomes i.e. their genetic material sometimes have an advantage over others”. Like the genome of other coronaviruses and unlike that of living beings, SARS-CoV-2 is entirely made of RNAFermerRibonucleic acid, which is chemically very similar to DNA (of which it is a sort of copy) that carries genetic information in the form of a long series of nucleotides (or “letters”).. A chain of four nucleotides – adenine (A), guanine (G), cytosine (C) and uracil (U) – this RNA can undergo mutationsFermerA random modification of genetic information on one, two or a small number of nucleotides (or “letters”) in a DNA or RNA sequence during its replication or “copy”. The more a virus proliferates, the more these mutations are therefore likely to appear., or in other words one or more of its nucleotides may be added, replaced or deleted in a random manner. To identify these variations, they are allocated a number that localises them on the genome and on the resulting proteins, as well as letters that indicate the type of modifications that have occurred.
Although the idea of a mutant virus may be frightening, in most cases such changes have no impact on its dissemination or severity. There is thus a French variant that has not spread elsewhere in Europe. Overall, several dozen may exist on different scales throughout the world. The situation becomes more complicated when the mutations modify the transmissibility or virulence of the virus, or even its susceptibility to treatments and vaccines. “In our jargon as virologists, we only refer to variants when the mutations affect the antigenic response of the virus,” Bruno Lina adds. “The antibodies that reacted with the original agent are no longer as effective, or become ineffective when it comes to the variant. So at present, although the immune response induced by the original virus is still adapted to the ‘UK variant’, it appears to be less so regarding the Brazilian and South African ones.”
Mutant and opportunistic
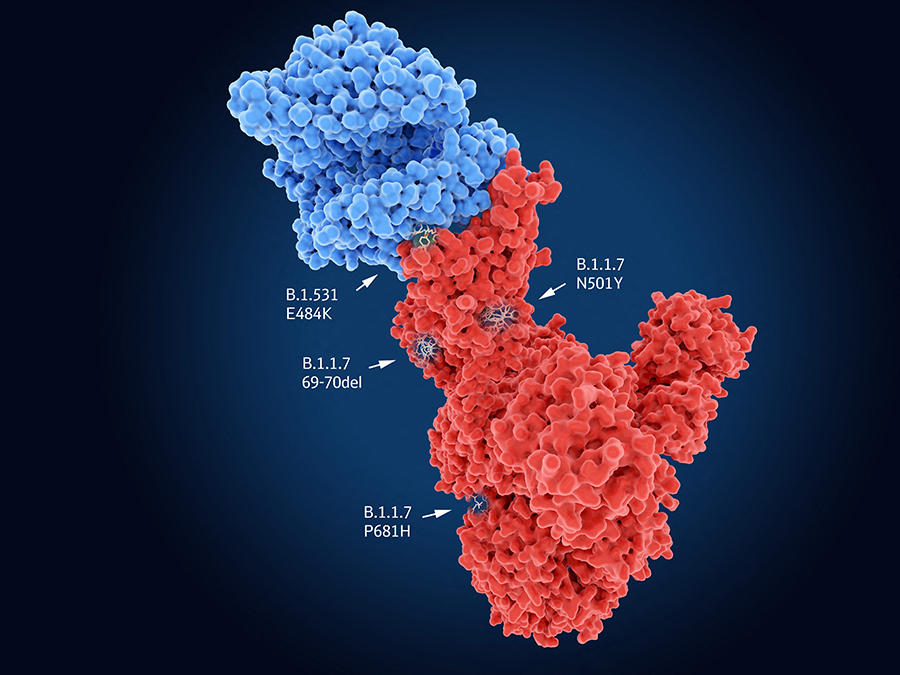
Like all viruses, SARS-CoV-2 requires cells to proliferate and spread. It therefore needs to enter its target in order to hijack the cell machinery to its own benefit. In the case of SARS-CoV-2, the Spike proteinFermerA spike-shaped protein found on the surface of the Covid-19 virus that enables it to penetrate into the cells it wants to infect. recognises cell receptors before penetrating into its host. Indeed, the UK variant carries the N501Y mutation on this protein, which thus facilitates entry of the virus into the cell and may render it more contagious. The genomes of the South African and Brazilian variants also contain N501Y, but combine it with the E484K mutation that neutralises part of the response of individuals immunised against the original lineages of SARS-CoV-2. Yet these common features are not necessarily bad news…
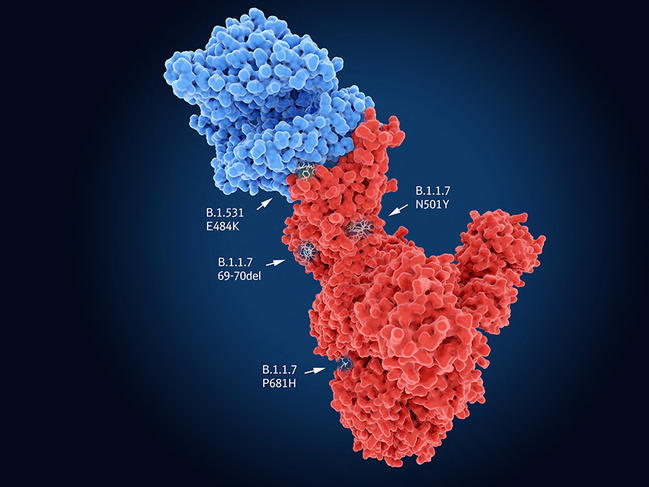
Indeed, “although the mutations thus converge towards the same evolutionary hot spots – which the monitoring of different lineages is foreshadowing – the virus may ultimately find itself in a dead end; it will no longer have sufficient room for manoeuvre to produce new variants and continue to adapt to humans”, suggests Bruno Lina. “In this way, the situation may ultimately stabilise.” This status quo might be welcome in view of the announced efficacy of the vaccines already developed. Less optimistic is Samuel Alizon, a CNRS senior researcher at MIVEGEC2 and a member of the modelling group within the laboratory’s Evolution Theory and Experiments (ETE) team, who believes that “the range of possibilities has changed because variants may continue to evolve by mutation in an as yet unknown adaptive landscape”.
An on-board safety net
What is the mutation capacity of the virus and where does it come from? “Influenza, dengue, Zika, chikungunya, AIDS, Covid-19, etc.; all viral pandemics in recent years have been caused by RNA viruses”, explains Isabelle Imbert, professor at Aix-Marseille Université and member of the AFMB3 laboratory. “They adapt more easily than DNA viruses because they do not repair the errors that appear during replication of their genome and thus mutate more frequently. However, coronaviruses are an exception to this rule: they are the only member of this family to be equipped with a corrective mechanism.”

Indeed, coronaviruses differ by the length of their genome; while the RNA virus of hepatitis C contains fewer than 10,000 nucleotides, that of SARS-CoV-2 has 30,000! With so many opportunities for mutations, the virus would disappear rapidly without its corrective “safety net”. Instead, it only generates two mutations per month on average, which is a hundred times less than HIV. This is why the genome of the UK variant displays only some 17 differences from that of the original strains that appeared more than 14 months ago. “In an infected cell, the viral genome is replicated thanks to a chemical polymerisationFermerA chemical reaction or process that enables the design of large molecules (containing a large number of atoms) by triggering reactions between smaller molecules. reaction during which copy errors may occur,” details Isabelle Imbert, who has published several studies on the subject. “If there is a problem, polymerisation is slowed thanks to two small viral proteins that enable the recruitment of exonuclease, an enzyme that is also coded by the virus. This can cut the RNA and ‘remove’ the incorrect nucleotide incorporated by mistake. This corrective system or ‘proofreading’ also exists in DNA organisms. Without this series of reactions, coronaviruses would mutate twenty times more rapidly.”
High rates of transmission
In the triple therapy used to treat HIV, one of the compounds does indeed target the polymerisation phase of the viral genome. The drug combination is able to attack the virus on several fronts, thus preventing it from mutating sufficiently quickly to defend itself against the three agents at the same time. Because some of the viral enzyme activities involved are not present naturally in our cells, the side effects are reduced. In the case of SARS-CoV-2, the replication complex is also a therapeutic target of choice alongside the Spike protein on which most vaccine strategies are based. “Numerous studies are focusing on the Spike protein with all its known mutations, and measuring the neutralisation of antibodies from vaccinated patients,” explains Isabelle Imbert. “In terms of the evolution of variants, so-called ‘RNA messenger’ vaccines have the advantage of being highly adaptable because it is simple to produce new RNA sequences coding for the Spike protein and its mutations. On the contrary, vaccines based on injecting the purified Spike protein i.e. once it has been isolated from the rest of the virus, as is the case with the Novavax vaccine are less reactive”.



According to figures published on 11 March by the French health agency, Santé Publique France, some 66% of people who tested positive were infected by the UK variant and 5% by the Brazilian and South African ones. Epidemiological models focus in particular on the reproduction number (R), or the number of people who are contaminated by each newly-infected patient. Rated R0 at the start of the pandemic (because it then corresponded to a constant or reference value) this number can vary over time (as a result of social distancing, for example) and is then rated Rt. When it is lower than 1, the epidemic recedes. Rt was around 0.9 in France before the emergence of variants, which means that 10% fewer people were infected during each cycle of infection. Yet with viral strains that are around 50% more contagious, Rt has once again risen above 1. Once these variants have replaced the original strains, there is a risk that the epidemic will flare up again. Protective measures are therefore needed to adapt to the situation, which unfortunately remains unprecedented.
Most outbreaks in history occurred when scientists and physicians had no genetic tools to monitor the lineages involved. Those caused by coronaviruses, such as the Severe Acute Respiratory Syndrome (SARS, which appeared in 2002 in China) and Middle East Respiratory Syndrome (MERS, seen in 2012 in Saudi Arabia) were rapidly contained because they did not involve many contaminations by asymptomatic individuals. The isolation of patients was sufficient to control the epidemic.
Viral modelling
“There is no well-documented example of the appearance of viral variants with such considerable respiratory transmission,” regrets Bruno Lina. “There are however similarities with chikungunya, which did not pose major problems when it was only being transmitted by a single type of mosquito, but suddenly spread through the western Indian Ocean when a series of mutations enabled it to infect more hosts, including urban mosquitoes.” The development of immunity finally put an end to the epidemic because these mutations did not affect the antigenic response of the chikungunya pathogen. On the other hand, changes to the influenza virus every year help it to re-infect individuals who have already been affected. Between these two extremes, it is still difficult to predict how SARS-CoV-2 will behave.
Distribution over time and in space
Epidemiological models shed light on the situation. “We started to work on the epidemic in March 2020 when we understood that the similarity of this virus to that of SARS had given rise to a false sense of security and the initial incidence figures had probably been vastly underestimated,” explains Mircea Sofonea, a lecturer at Université de Montpellier and member of MIVEGEC, who heads the Covid-19 epidemic modelling group4 set up by the ETE team. Models help to refine the data from screening, since the people tested are not necessarily representative of the overall population. All the figures therefore need to be smoothed and corrected in order to obtain an overview of the epidemic that will be as accurate as possible. “Our tools generate quantitative information, rather than intuitions or oral models,” insists the academic. For example, the working group has designed the Rt25 and COVIDici5 applications that enable real-time visualisation of the dynamics of the epidemic. “Without protective measures, the R0 of the original strains is 3, but now reaches 4 for the three variants that are currently making the headlines,” the scientist says. This means that each infected individual would contaminate another four, triggering an exponential spread of the disease. “The last release from lockdown in December 2020 took place while the circulation of the virus remained high, and the 6 pm curfew implemented throughout France in mid-January only froze this situation. But our role as scientists is to produce quantitative analyses, not advocate health policies.”
As well as its spatial distribution, the course of the virus is also studied over time. “Phylodynamics traces family trees of infections based on viral genomes,” explains Alizon. “In principle, SARS-CoV-2 originated from bats and passed to humans between August and November 2019. Apart from its acquisition of the D614G mutation, there was no massive spread of mutations conferring an advantage on the virus until the appearance of variants at the end of 2020.”

How have lineages carrying similar mutations emerged so close together in time and in several different places? The explanation is similar to the phenomenon of epistasis, which describes how genes interact with one another. Some mutations, such as N501Y which is present in the variants that have emerged in the UK, South Africa and Brazil, will only have a major impact if they are combined with a series of other mutations that are benign on their own.
Circumventing the immune response
“Although the risk of an isolated virus accumulating all the mutations that favour its spread is statistically very low, it is not negligible when several millions of people are infected,” warns Alizon. “Furthermore, the immunisation of populations modifies selection pressures.” The scientist cites the example of the city of Manaus in Brazil, where the authorities allowed the virus to spread. This approach not only caused a health disaster in 2020, but it might also have led to the evolution of a variant that was able to re-contaminate some people despite their hard-won immunity against the original strain. Another hypothesis that explains the emergence of variants is that of immunosuppressed patients, in whom it is known that viruses can replicate for several months thanks to the individual’s weakened immune response. What is not known though is whether the variants have altered viral virulence, i.e. the ability of the virus to proliferate rapidly and increase the patient’s viral load, which in turn can trigger severe forms of the infection.
With respect to the number of deaths caused, a study published on 10 March in the British Medical Journal showed that the UK variant was 64% more lethal in symptomatic patients than the original strains.6 And modelling performed at the end of December 2020 and published on 3 March in Science showed that this variant, which is much more contagious than previous lineages, could result in higher fatality rates at a population level because it causes more infections.7 “With the number of deaths we have suffered, we would already have reached herd immunity if the virus causing Covid-19 had been as virulent as the influenza pathogen. Sadly, it is ten times more infective,” deplores Alizon. “There is no data at this stage to determine whether variants will become increasingly virulent, but their high contagiousness may lead to a rise in death rates. Epidemiological models suggest that once the population is immunised, outbreaks caused by this virus could prove far less lethal and more similar to the seasonal coronaviruses that infect us every winter. Yet it is too early to know whether and when this will happen, particularly in view of the lack of knowledge on the capacity of variants to escape the immune response while remaining virulent.”
- 1. CNRS / Université Claude-Bernard Lyon 1 / INSERM / ENS de Lyon.
- 2. Laboratoire Maladies Infectieuses et Vecteurs: Écologie, Génétique, Évolution et Contrôle (CNRS / IRD / Université de Montpellier).
- 3. Architecture et Fonction des Macromolécules Biologiques (CNRS / Aix-Marseille Université).
- 4. https://covid-ete.ouvaton.org/
- 5. https://cloudapps.france-bioinformatique.fr/covidici/
- 6. https://www.bmj.com/content/372/bmj.n579
- 7. https://science.sciencemag.org/content/early/2021/03/03/ science.abg3055
Explore more
Author
A graduate from the School of Journalism in Lille, Martin Koppe has worked for a number of publications including Dossiers d’archéologie, Science et Vie Junior and La Recherche, as well the website Maxisciences.com. He also holds degrees in art history, archaeometry, and epistemology.