You are here
The evolution of the Covid-19 virus remains unpredictable
The Omicron wave seems to have passed and the war in Ukraine has taken over the headlines. But where do we actually stand regarding the Covid-19 epidemic and its variants?
Samuel Alizon:1 Our perception of the epidemic is too easily influenced by its trends in France: the picture looks dire when numbers rise and seems rosy when they fall. This reaction is even observed amongst researchers, and should be taken with caution. At the international level, the situation is clearly not synchronised, in particular with a wave of the Omicron variant currently causing numerous deaths in South-East Asia. We should therefore bear in mind that pandemics are a long-term phenomenon and involve marked local differences. In other words, we should not extrapolate too much from the current context in a given country.

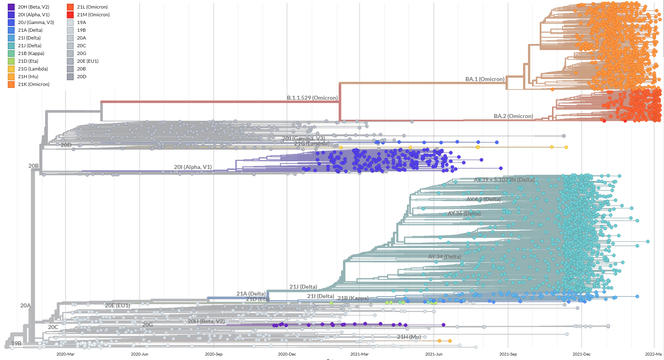
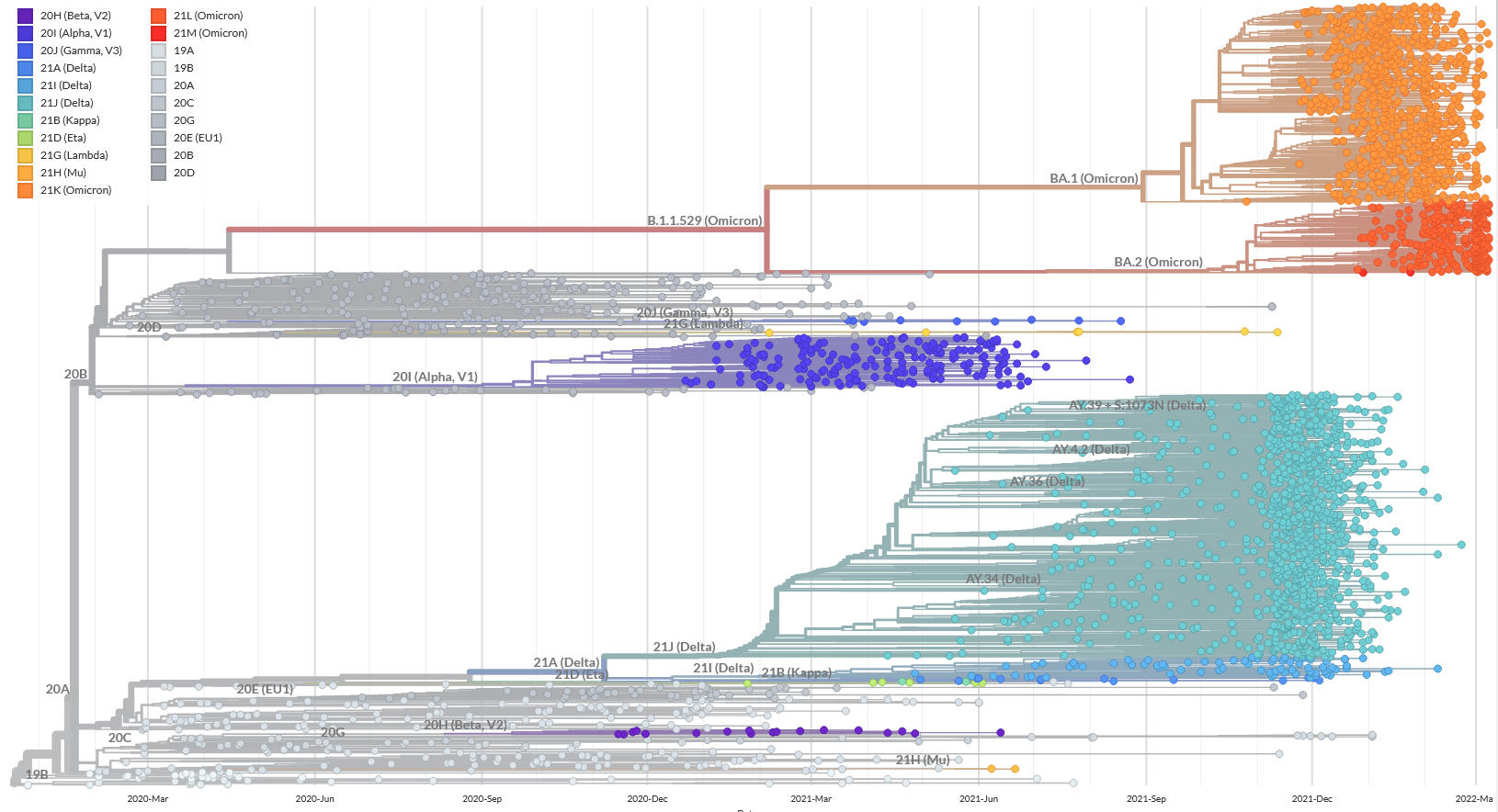
That said, at present (this interview took place on 8 March; Editor’s note), the BA.2 variant is clearly dominant in France. It is a sister to BA.1, the Omicron strain that caused the wave in January. Both of them probably diverged about a year ago in southern Africa. BA.2 differs as much from BA.1 as the Delta variant differed from Alpha, but shares some of its characteristics: it is about twice as contagious as Delta and three times less virulent in a vaccinated population. The reasons for this replacement have been debated, but according to our British colleagues, it may be a matter of temporality, as BA.2 infections become contagious slightly more rapidly. However, we still do not know the extent of the epidemic peak, because alongside biology, national policies and individual behaviours also play an important role.
Although some underlying trends are becoming apparent, the progression of variants remains largely unpredictable. The Alpha strain appeared when most epidemiologists thought that the evolution of SARS-CoV-2 was only neutral. At a time when there were fears that mutants of Alpha might surface and escape immunity, Delta emerged and replaced it. And when monitoring focused on Delta clusters carrying worrying mutations, Omicron took over even more rapidly. Nevertheless, it marked a turning point, raising questions as to whether we are not experiencing progressive branching, or in other words a switch from one to two different lineages.
Indeed, Omicron viruses appear to exploit an ecological niche which differs from that of other SARS-CoV-2 variants, i.e. the lower respiratory tract. Instead, Omicron tends to affect the upper respiratory tract where the immune response is less strong. We do not know how long these two types of variant will coexist, each with its preferred target in our bodies, or whether BA.2 will take over.



What are the possibilities described by research?
S. A.: The UK, which is by far the most advanced in addressing these questions, issued a report in February this year containing four scenarios regarding the long-term evolution of the epidemic, with a timeline of more than 18 months. One is optimistic, two are considered as average and the fourth is pessimistic. The first projection corresponds to a continuation of the current situation in Europe, where the virus is displaying low immune escape and the severity of infection is limited thanks to vaccination and treatments. Although winter waves remain possible in this context, their scope will be limited, and booster jabs will only concern people at risk.
Immune escape would be greater under the average scenarios, causing further severe new waves, potentially as soon as the autumn. We still do not fully understand the properties of long-term immunity, but once again, the British data indicate that protection is less durable against Omicron than against Delta. As a result, these scenarios could require further general vaccination campaigns in certain years. Finally, under the worst-case scenario, not only immunity would be low but the infection would be more virulent, if only because of the emergence of a new variant or the generalisation of viral resistance to treatments. It would then be necessary to update the composition of vaccines or turn to non-pharmaceutical interventions. At present it is difficult to make projections with any certainty for more than a few months, and we are limiting ourselves to determining the most likely scenarios. In light of the modelling performed by our team for the medium term, both the optimistic and pessimistic options seem unlikely, but once again, the variants have shown that rare events can have global consequences.
How is the monitoring of variants organised?
S. A.: Once again, our British colleagues have an unquestionable advance over us in terms of the quality of their surveillance and analysis. But monitoring the appearance of new variants requires excellent knowledge in the field. At the international level, this therefore means trusting epidemiologists in each country. This is what happened with Omicron in South Africa, where scientists shared their concerns with the rest of the world when they saw that the epidemic was displaying local specificities in several provinces and was no longer behaving in the same way.
Typically, the detection and monitoring of variants of concern are based on three criteria: epidemiological, pathological and genetic. As a general rule, localised clusters or outbreaks of an epidemic are observed that run contrary to the underlying trend. Field studies coupled with the sequencing of viral genomes then show that these upturns are caused by a specific strain that gives rise to different symptoms in terms of immune escape, contagiousness or virulence. France rapidly implemented an epidemiological network to monitor these criteria, but compared to the UK, we are in the position of spectators, especially when it comes to the analysis of variations in severity or immune escape. However, it should be pointed out that British law is much less strict than ours on the use of medical data, which does raise some difficult ethical questions. Furthermore, for many years, English-speaking countries have established good synergistic relationships between academic research and public health organisations.
From a genetic standpoint, France was one of the first countries to sequence the genome of SARS-CoV-2, the virus that causes Covid-19, but then fell behind. The Emergen project,2 coordinated by Public Health France and ANRS-MIE,3 restored the balance in terms of the quantity of genomes produced, but we lagged far behind the UK with regard to data quality and exploitation. France also adopted an approach based on genetic screening: rather than sequencing the entire viral genome, this technique only detects a few specific mutations in variants. This enables the testing of all positive samples, which would be much too lengthy and costly with the sequencing of complete genomes.
Our team was thus able to issue rapid alerts regarding the waves of the Alpha, Delta or Omicron variants. Such exhaustiveness and reactivity was at the expense of specificity; for example, in France, it is still impossible to distinguish BA.1 from BA.2 by means of screening.
Before Omicron emerged, some people had expressed their hopes that the Covid-19 virus had already exhausted a large proportion of its evolutionary potential. How is that measured and where do we stand at present?
S. A.: We did indeed find several mutations in common between the different variants. The most striking case was the D614G mutation which appeared very rapidly and independently in different countries. Such cases of parallel evolution are generally the sign of strong selection pressure. However, the selective advantage provided by mutations does not necessarily mean fewer possibilities. On the contrary, a mutation may open up a new evolving landscape, as appears to have been the case with the N501Y mutation. In defence of the hypothesis of progressive narrowing, the organisation of viral genomes imposes enormous constraints in mutational terms. While our DNA is made up of several billion base pairs, SARS-CoV-2 must complete its entire life cycle with 29,000 pairs, in other words enter an organism, duplicate its genetic material in its cells and produce new virions, all while escaping the immune response. For this reason, most of the possible mutations have a deleterious effect on the virus; some are neutral and only a minority can be beneficial.
Mutagenesis experiments are able to artificially mutate certain parts of the pathogen in order to study any phenotypic changes; i.e. affecting their properties. Of course, this is performed on fragments and isolated proteins, and not on a complete and viable virus. Work carried out so far has mainly focused on the Spike protein, which lies at the interface between viral particles and our cells. Such studies, and notably those conducted by Jesse Bloom’s team in the Fred Hutchison laboratory,4 have thus identified mutations that enhance several hundred-fold the affinity of this Spike protein for the ACE2 receptor, which serves as a portal of entry for the virus into a cell.
But although it is possible to anticipate the effects of a mutation taken in isolation, the dimension of potential combinations renders this operation almost impossible when several mutations need to be considered. Indeed, the Omicron variant already differs from the ancestral Wuhan strain by more than fifty mutations.
Are there any means to limit this mutation potential and hence the emergence of new variants?
S. A.: Each time someone is contaminated, billions of new viral particles are produced, and as a result of copy errors, increase the risk of emergence of random mutants. Because of recombination, this effect is enhanced when a person is infected by several different strains: if two viruses are present in the same cell, exchanges of whole portions of their genomes can occur. Once again, mutations and recombinations are very likely to be harmful to the virus, but some may also be a problem for us. And with enormous population sizes, the unlikely can become frequent. However, to summarise, the greater the increase in viral circulation, the more its evolution will accelerate. In this regard, documented cases of the spread of SARS-CoV-2 in animal reservoirs, even in the New York sewers, are a cause for concern.

Finally, to understand viral evolution, we need to look at intra-host dynamics. In most people, the infection lasts less than two weeks and it is estimated that a new one is only established by one or three virions. This bottleneck markedly limits diversification of the virus. In a way, viral diversity is practically back to zero at each new contamination. Nevertheless, in immunocompromised individuals, the infection can last for several months and cause severe complications. From an evolutionary point of view, the process is no longer disrupted by weekly bottlenecks and viral diversity increases over the months. This boosts the rate of evolution because mutations drive natural selection. Several studies have thus shown that some strains that evolve during infections in immunosuppressed patients carry key mutations of certain variants. The essential prevention and monitoring of immunocompromised people must therefore be both health-based and evolutionary. In any case, the implementation of preventive policies in order to reduce the number of contaminations to a minimum is the best way to prevent the risk that new variants of SARS-CoV-2 will appear.
- 1. CNRS Research professor at the Centre for Interdisciplinary Research in Biology (CIRB – CNRS / Collège de France / INSERM).
- 2. Consortium for surveillance and research on emerging pathogenic infections through microbial genomics.
- 3. French Agency for Research on AIDS and Viral Hepatitis – Emerging Infectious Diseases (ANRS-MIE).
- 4. Fred Hutchinson Cancer Research Center (Seattle, USA). Captions
Explore more
Author
A graduate from the School of Journalism in Lille, Martin Koppe has worked for a number of publications including Dossiers d’archéologie, Science et Vie Junior and La Recherche, as well the website Maxisciences.com. He also holds degrees in art history, archaeometry, and epistemology.