You are here
Bioinformatics, a key ally in the fight against Covid-19

“The Covid-19 pandemic has been a real-life test for bioinformatics. It has revealed how the tools we are developing have become essential for analysing data and accelerating research on the virus.” According to Hélène Touzet, at the Research Centre in Computer Science, Signal and Automatics of Lille (CRIStAL),1 who coordinated a recent report on the subject,2 the outcome is very clear: if in such a short time scientists have been able to uncover numerous secrets about SARS-CoV-2, ranging from its identification to its origin, and including its functioning and spread, this is largely due to bioinformatics – or computational biology. At the crossroads between information technology, mathematics, biology and physics, this recent discipline – it is only about twenty years old – provides calculation models, algorithms and other software programs that can process an avalanche of biological data and discover key information.
Sequencing SARS-CoV2
Bioinformatics proved its worth right from the start of the pandemic. After identifying the first individuals infected by the virus in December 2019, Chinese scientists immediately set out to sequence the genome – genetic material made up of RNA, a long chain of nucleotides – of the new pathogen using samples collected from these patients. And although this operation was actually performed by biologists, it was bioinformatics algorithms that enabled them to produce the full sequence.

The raw data obtained is in fact “illegible”: it is made up of millions of small, partial sequences of several hundred nucleotides from diverse origins; they may be pieces of the viral genome but can also arise from the patient or even bacteria. To clarify these findings, an initial algorithm compares all these fragments with those present in a massive database that contains all genomes from the living world listed to date. In the same way as a Google search on the internet, all unknown sequences are thus identified, which means they belong to the new virus. A second algorithm, based on pathway optimisation methods in networks, then puts all the pieces together in the correct order.
Comprising around 30,000 nucleotides, the SARS-CoV-2 genome was thus revealed in record time – only twelve days! “This was made possible thanks to both advances in algorithms over the past ten years and to open source access to software programs, as well as the global sharing of genomic data. This trend started during the 2010s, especially in the context of the Ebola epidemic, but has now become the norm with Covid-19,” Touzet notes.
Establishing the genealogy of the virus
Once the genome of the virus was established, bioinformatics once again helped to derive valuable information from it, first of all about its origin. By comparing the new genetic material with that of other known coronaviruses, the researchers demonstrated that SARS-CoV-2 displayed greater similarity to pathogens present in bats than to the human SARS and MERS viruses that had respectively caused the epidemics in 2003 and 2012. This strongly suggests that SARS-CoV-2 originated from these mammals – probably with another host animal acting as an intermediary between this species and humans.

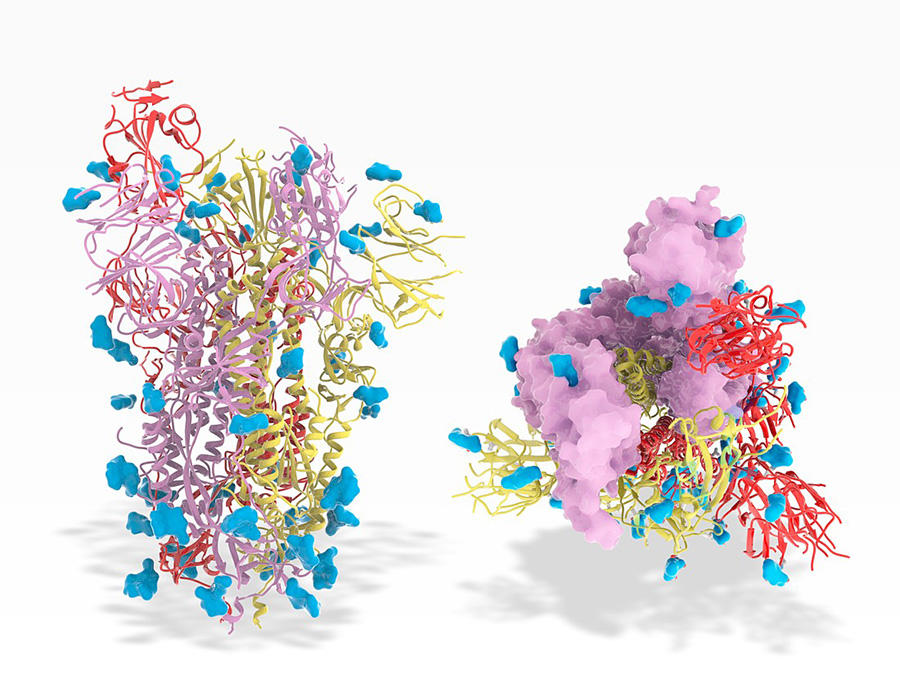
The comparative analysis of genomes by algorithms also provided scientists with a first insight into the biology of the virus. “By analogy with other pathogens for which we had already produced a photofit in the laboratory, it was possible to identify sequences that coded for proteins and determine the latter precisely, amino acid by amino acid, in order to compare them with those in other viruses and highlight common features and differences,” the scientist explains. The biologists were thus able to discover the principal proteins required for the virus to function – like all pathogens, SARS-CoV-2 needs a host cell to multiply and spread – and particularly those responsible for infecting human cells. They included the famous Spike protein, which recognises cell receptors before penetrating the host, and is now the focus of biologists’ attention since it is the target for the Covid-19 vaccines currently on the market.
Modelling molecular mechanisms
Although it is of crucial importance, genomic data alone is not sufficient to elucidate how the virus functions once it has entered the human body. Yet to fight the pathogen, biologists must be able to describe in detail the molecular mechanisms at play. To achieve this, they rely on structural bioinformatics, a specific field of bioinformatics. Thanks to digital simulations that can model all the actors involved at the atomic scale, the scientists seek to determine the three-dimensional structure of molecules and understand how it affects their interactions.
In the case of SARS-CoV-2, such models have provided support for experiments (crystallography, electron cryomicroscopy, etc.) aimed at identifying the structure of the Spike protein and shedding light on its docking dynamic with a host cell. This proved crucial for the development of vaccines.
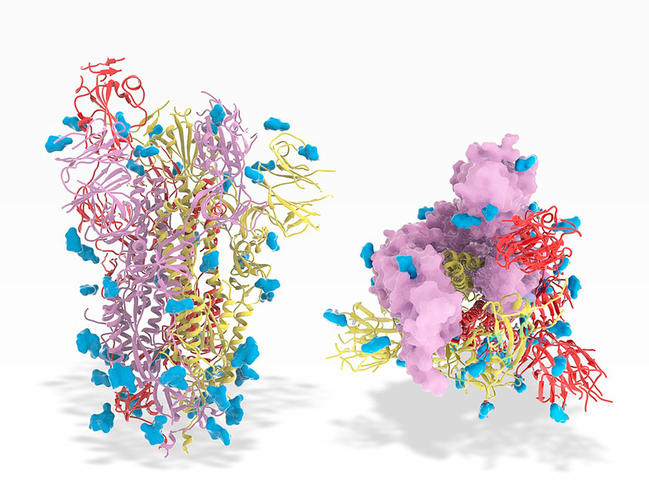
Such structural modelling can also help to predict how a mutation – i.e. a modification to the RNA sequence of the virus that occurs randomly when the pathogen reproduces – can change the shape, and hence the function, of a protein; for example by replacing one amino acid by another. In the case of SARS-CoV-2, biologists thus keep a watchful eye on any mutations that might affect the Spike protein and even render a vaccine strategy ineffective. “One of the ultimate ambitions of bioinformatics would be to simply predict the structure of a protein from the genome sequence. Much work remains to be done to achieve this, but with our existing tools we can already estimate the potential impact of mutations on the conformation of a protein and hence the risk of vaccine resistance,” Touzet explains.
Monitoring the evolution of the virus
Bioinformatics has other advantages. As well as revealing the origin of the virus and its biology, genomic data – once again – can retrace how an epidemic has spread. “The regular accumulation of mutations in the genomes of viruses during a period of infection – on average one or two per month – can be used as a sort of ‘molecular clock’. If people are contaminated by similar viruses, this means that they are close in the chain of transmission. Comparing the genetic material of viruses in infected patients, and using information on the dates and sites of sampling makes it possible to trace the evolution of an epidemic over time and space,” explains Samuel Alizon from the MIVEGEC laboratory.3
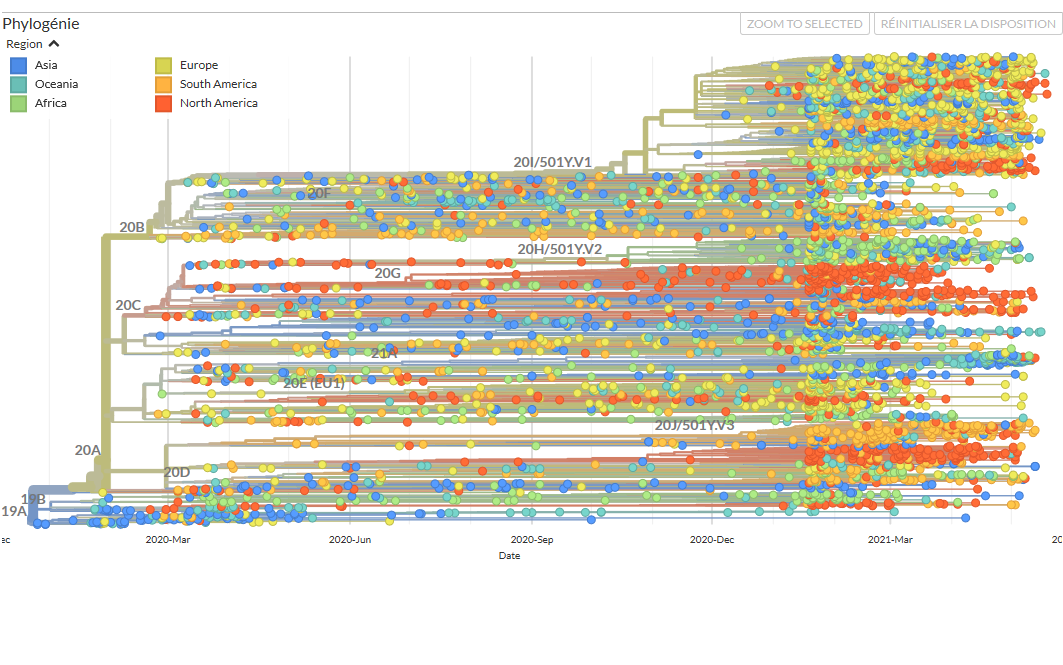
The emerging discipline of phylodynamics is experiencing substantial growth in the context of Covid-19. In practice, bioinformatics specialists use genomic data to build a phylogenetic tree, or a family tree that links the different infections in which the viruses have been sequenced. To do so, they apply the statistical principle of maximum probability: the tree thus established is that which best explains the relationships between the sequences available.
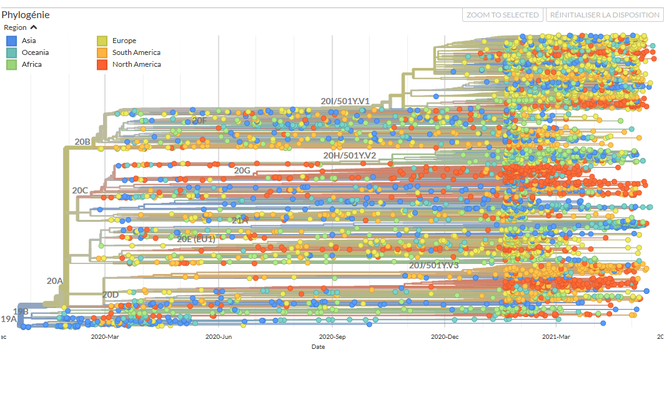
An enormous quantity of information can be drawn from these trees, in particular regarding the origin of the pandemic. Thus by comparing some fifty viral sequences collected from Chinese patients at the start of the outbreak, scientists observed a weak genetic diversity of the virus. This suggested that it had appeared only recently – between August and December 2019 – in the human population, following a single contamination by an animal – and that no more transmissions subsequently occurred between animals and humans.
Elucidating the origins of the pandemic
These genealogical trees have also informed the history of the pandemic, country by country. For example in France, an analysis by scientists, including Alizon, of each genomic sequence available in March 2020 revealed that they all descended from a single common ancestor dating from the early days of the oubreak in China. The fact that several viral lineages from that country were found in France suggests that separate “imports” of the virus contributed to the first wave of the epidemic. In the UK, where many more sequences are available, this number has been estimated at more than a thousand.
Another illustration, this time at the scale of a city, is Boston, in the US. American scientists showed that an international trade conference in February 2020 was a “superspreader” event which, according to their estimates, probably led indirectly to more than 100,000 contaminations throughout the world over the following nine months. These findings are crucial to investigate the causes of such events and prevent them in the future.
“Phylodynamics is an invaluable tool to monitor an epidemic, particularly since the data it uses offers the advantage of both minimising certain analytical biases, compared with incidence data the number of new cases by time unit – editor’s note arising from screening – and ethical risks, as opposed to digital tracing data,” suggests Alizon. With his team, the researcher is hoping to go even further by combining genomic and traditional epidemiological data, thus greatly improving the overall monitoring of the virus.
- 1. CNRS / Université de Lille / Centrale Lille.
- 2. Available at https://hal-cnrs.archives-ouvertes.fr/hal-03170023.
- 3. Maladies Infectieuses et Vecteurs: Écologie, Génétique, Évolution et Contrôle (CNRS / IRD / Université de Montpellier).
Explore more
Author
A graduate from the School of Journalism in Lille, Martin Koppe has worked for a number of publications including Dossiers d’archéologie, Science et Vie Junior and La Recherche, as well the website Maxisciences.com. He also holds degrees in art history, archaeometry, and epistemology.