You are here
A Biomolecular Kaleidoscope
An international collaboration1 bringing together French and Japanese computer scientists, engineers, physicists and chemists—led by the LIMMS2 CNRS/University of Tokyo unit—has just developed a valuable new tool for cracking the secrets of biomolecular processes: a device that can test the biochemical reactions of a given set of components in around ten thousand different types of conditionsFermerUsing different proportions of substances or compounds (reagents) to test whether or not a chemical reaction occurs.—simultaneously.3
A microfluidics revolution
At the root of this system is microfluidics, a multidisciplinary field that emerged in the early 1980s focused on the manipulation of tiny volumes of liquids (think dozens of picoliters, the volume of a large human cell) and which is already used for inkjet printheads, lab-on-a-chip devices or next-generation sequencing.
Using microfluidics, the researchers were able to significantly upgrade existing reaction-testing methods, notably by reducing testing time from weeks or months to a matter of days. And this comes as good news for the medical field in particular, as this technique could enhance molecular diagnostics for a wide range of illnesses.
10,000 windows
To better understand chemical processes in living organisms, but also to engineer molecular solutions for various purposes (such as diagnostics), scientists need to understand how a set of reagents will react under a broad range of conditions, i.e., with different concentrations of reagents. For now, such reactions are investigated via computer simulations or laboratory experiments—both of which present drawbacks. Simulations, which often rely on educated guesswork on how molecules behave, may fail to convey the full scope of reality, while hands-on experimentation, even for simple setups, is time-consuming and lengthy (up to months). The new Franco-Japanese technique is able to overcome these drawbacks, churning out ten thousand or so experimental data points, while also slashing the time and effort required for preparation thanks to the minuscule volumes involved in microfluidics, providing results in a matter of days.
This remarkable innovation marks more than two decades of French-Japanese scientific collaboration at the LIMMS, the project’s key research unit. Launched on the Komaba campus of the University of Tokyo in 1995, it has so far hosted over 150 French and other European scientists, matching them with Japanese partners. “Regarding this particular project, researchers on the French side—experts in DNA nanotechnologies or molecular engineering—knew little about the possibilities of microfluidics, whereas their Japanese counterparts, microfluidics specialists, were not intimately acquainted with the engineering challenges of our field, namely the need to test a huge number of experimental conditions to get a system to work,” explains LIMMS researcher Anthony Genot, who believes the international and multidisciplinary atmosphere fostered by the unit played a key role in developing this device.
“Our main objective here is to program biochemical networks by drawing inspiration from what occurs in cells,” adds fellow LIMMS researcher Yannick Rondelez. “But even if biochemical networks can compute like electronic circuits, we often don’t know exactly how their chemistry works, so we need to test a great variety of experimental conditions.” A pooling of French and Japanese know-how thus helped to build the new device, which now brings to molecular engineering “the many benefits of microfluidics—which include low volume, automation, parallelism, and speed of preparation,” says Rondelez.
A small platform with big prospects
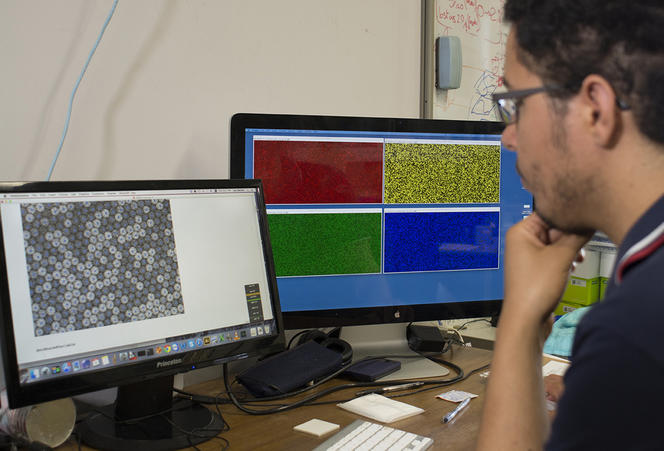
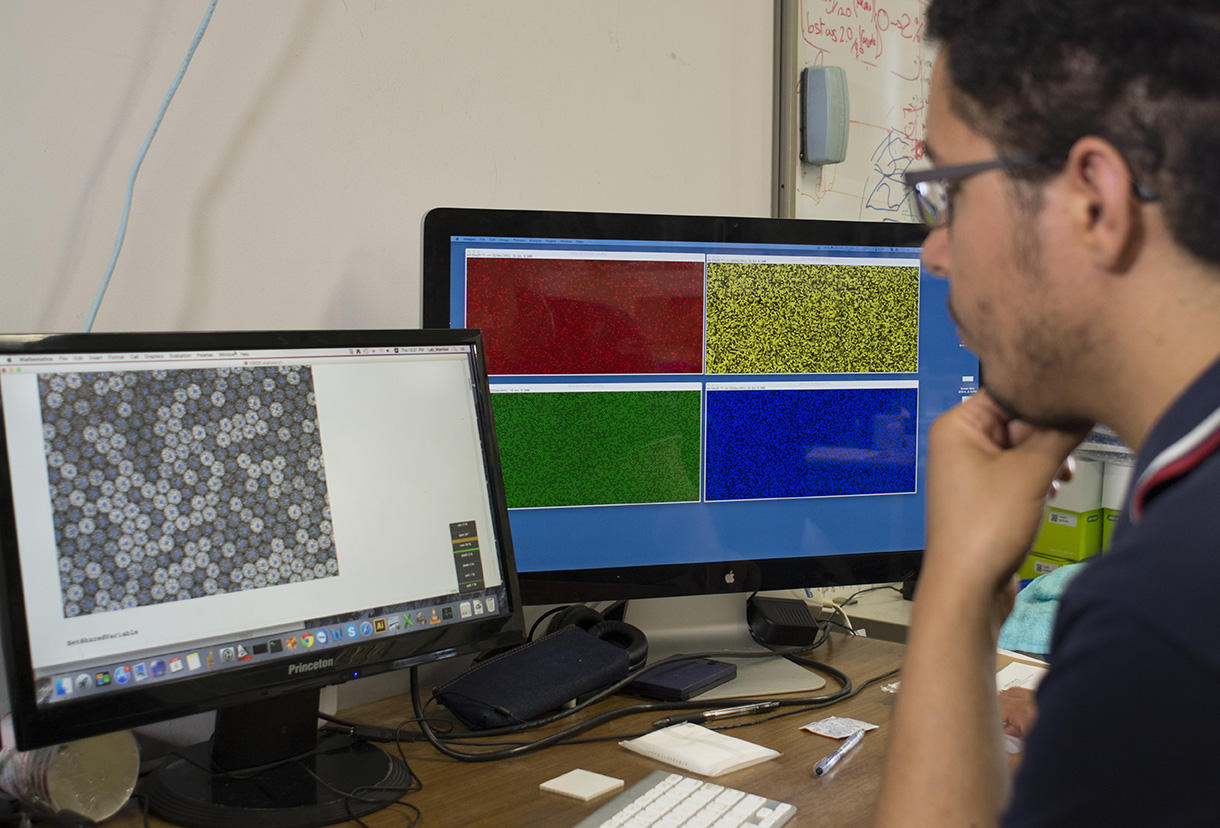
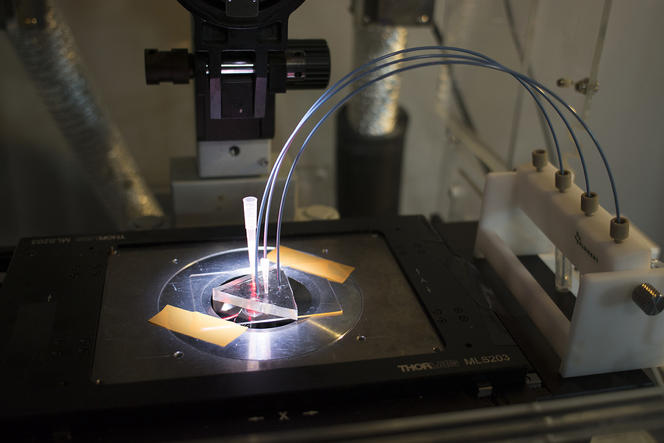
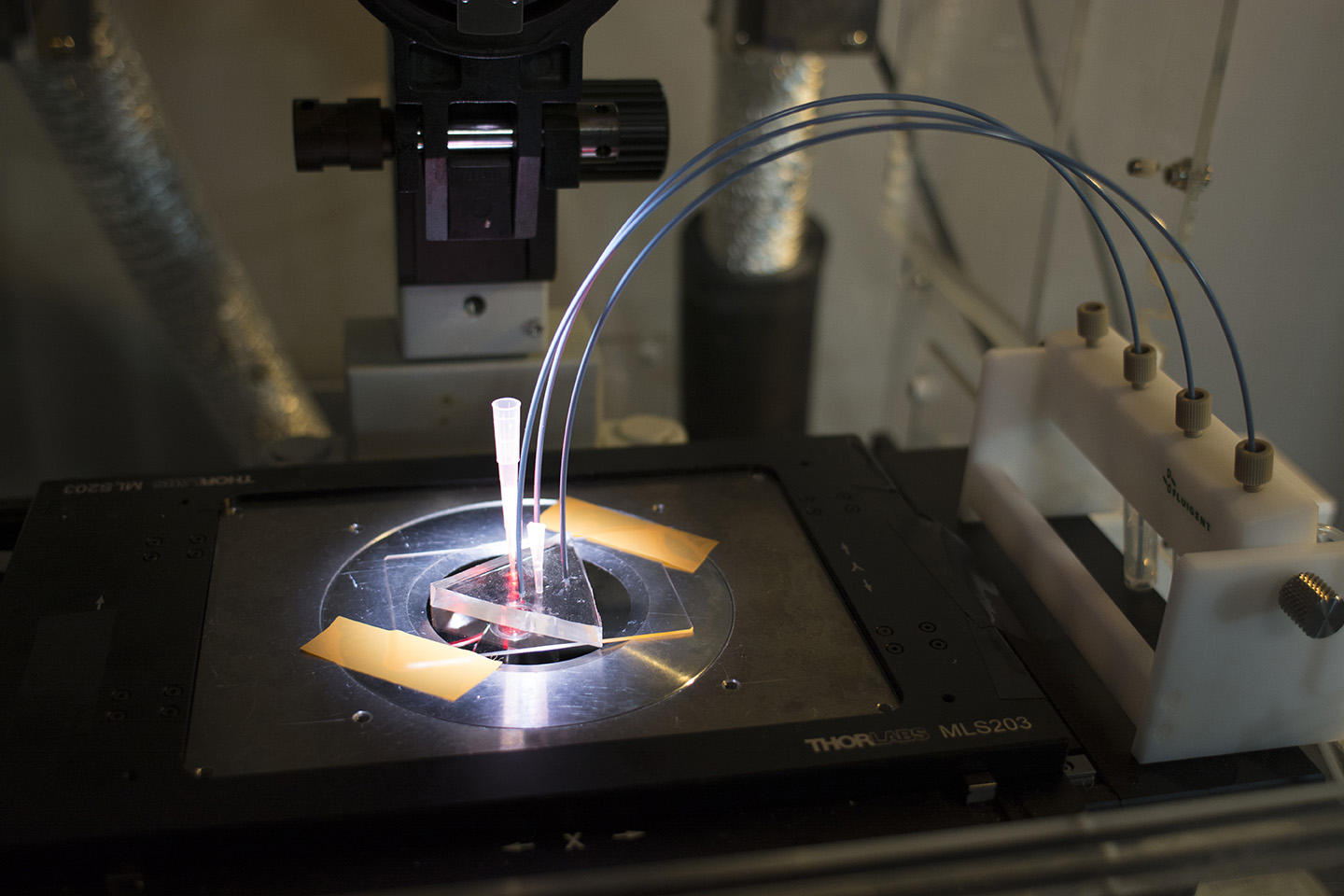
Concretely, the device—elaborated in the Applied Microfluidic Systems Laboratory (led by Teruo Fujii) at the Institute of Industrial Science in Tokyo—consists in a platform that generates a myriad of uniform, micrometer-sized droplets, each containing random concentrations of given reagents. After sandwiching and immobilizing a single layer of droplets between glass slides, a microscope reads fluorescent markers associated with the reagents, revealing the chemical conditions in each droplet. The microscope images paint a colorful array of dots that progressively metamorphose—like a kaleidoscope—as reactions individually unfold in droplets.
Not only does the device yield high-resolution maps that inform on the dynamics of reactions and their optimal conditions, it also reveals experimental subtleties so far only hypothesized through computer simulations. “Furthermore, since we tested highly nonlinear systems that are some of the most complex we can create, this method should also apply to most biochemical protocols, whose behavior is mathematically far simpler,” Genot specifies.
For now, the versatile system has proven its capacity to chart reactions in ten thousand droplets at a time—a far higher throughput than offered by today's existing protocols able to collect around ten to one hundred data points per day. Ten thousand may already seem dizzying enough, but researchers aim to multiply the figure many times over in the future. The team’s initial experiments relied on a confocal microscope that provides high-quality images but operates relatively slowly. By migrating to a quicker microscope, Genot expects that the system could “measure hundreds of thousands, or even a million reactions at once.”
Among potential applications, the team foresees the optimization of enzymatic reactions for molecular diagnostics. This is the case for the ubiquitous PCR (polymerase chain reaction) method that amplifies traces of DNA to detect the presence of viruses, bacteria or certain types of cancers from biopsies or blood samples. “Whenever the diagnosis of a cancer or another illness requires a complex chemical protocol, such as the simultaneous detection of several DNAs, our method can contribute to improving the procedure,” Genot explains.
Apart from diagnostics, Rondelez adds that the device also accommodates a current trend in biotechnological and medical fields—in genomics for example—to have recourse to “increasingly complicated enzyme cocktails to manipulate and analyze DNA. The development of these systems requires the simultaneous balancing of several reagent concentrations—which is precisely what our device can do.”
As they continue to explore the wide potential of this device in high-throughput enzymology, the French-Japanese team will now also be turning its attention to streamlining the platform, namely by simplifying its use and upping its capacity to the million-droplet mark. Now that the LIMMS has harnessed microfluidics for the detailed mapping of biochemical processes, hopefully it won’t be too long until diagnostics improved by their high-speed technique reach a medical center near you.
- 1. Electrical Engineering and Applied Physics department (EEA), ENS Cachan; Earth-Life Science Institute (ELSI), Tokyo Institute of Technology; Institut des Systèmes Intelligents et de Robotique (ISIR, CNRS / Université Pierre et Marie Curie); Laboratoire de Chimie & Biochimie Pharmacologiques et Toxicologiques (LCBPT, CNRS / Université Paris Descartes); Laboratoire de Conception et Application de Molécules Bioactives (LCAMB, CNRS / Université de Strasbourg); Laboratoire Gulliver (CNRS / ESPCI Paris Tech); Laboratory for Integrated Micro-Mechatronic Systems (LIMMS, CNRS / Institute of Industrial Science / University of Tokyo); Laboratory for Analysis and Architecture of Systems (LAAS-CNRS); Ochanomizu University; Translational Research and Microfluidics (INSERM UMR-S1147 / CNRS SNC5014 / Université Paris Sorbonne Cité / Centre Universitaire des Saints-Pères); Université Pierre et Marie Curie, Sorbonne Universités.
- 2. Laboratory for Integrated Micro-Mechatronic Systems (LIMMS, CNRS / Institute of Industrial Science / University of Tokyo).
- 3. A.J. Genot et al., “High-resolution mapping of bifurcations in nonlinear biochemical circuits,” Nature Chemistry, 2016. DOI: 10.1038/NCHEM.2544.
Explore more
Author
As well as contributing to the CNRSNews, Fui Lee Luk is a freelance translator for various publishing houses and websites. She has a PhD in French literature (Paris III / University of Sydney).