You are here
Single-cell technologies mark the dawn of a new era
At the Institut Curie, a tumour specimen circulates from floor to floor. First collected from a patient by a physician in the basement, it then moves up to the pathology department to be prepared for study. It is analysed, cell by cell, using a revolutionary “single-cell” technology, before going up another floor for sequencing. Finally, the bioinformatics specialists clean the dataset produced by the sequencer and apply statistical methods to enable its study. The aim is to understand why a tumour has emerged and is resistant to treatment. Within a few months, the living specimen has become a body of data. “Single-cell methods have accelerated biology 2.0, and our work is now based on a multidisciplinary ballet from the physician to the biologist, the data analyst and statistician. It is only through the participation of all these specialists that the whole system works”, explains Céline Vallot, CNRS research professor at the Institut Curie1 and specialist in the epigenetic mechanisms of breast cancer.
Since 2017, Vallot and her team have been using and developing single-cell techniques for their research on cancer. This technology is an innovation that uses a process called microfluidics: one by one, the cells pass through micro-channels to become encapsulated in a micro-droplet of oil containing reagents. At the time of encapsulation, the genetic material in the cell is labelled using a genetic barcode that identifies the cell. This method enables the scientist to work at the scale of the individual cell and try to understand the cellular heterogeneity present within a single tumour. “Before single-cell technology, we performed experiments on groups of millions of cells and then studied the mean expression of the genes involved,” she explains.
Nevertheless, the epigenetics expert points out that the scale of a single cell is nothing new in biology. “When studying proteins, we were using microscopy methods. What is innovative with single-cell technology is its systematic nature, as we can focus on numerous cells and obtain relatively exhaustive information,” she adds. This new technique offers much hope for cancer research, and medicine in general, with one major opportunity: to create precision medicine where each patient receives a tailor-made treatment.
Multiple approaches
The first scientific publication2 in which the single-cell method was applied to study RNA dates from 2009. The technology gained momentum in 2015, and from 2017 most biology laboratories studying cancer in France were using it. At the Nantes-Angers Cancer and Immunology Research Centre (CRCI2NA)3 in western France, the equipment was introduced in 2018. No larger than a mini-dishwasher, it offered a myriad of possibilities for the team led by Stéphane Minvielle, CNRS research professor and jointly responsible for the Integrated Cancer Genomics (ICAGEN) team. For the past twenty years, this group based within Nantes University Hospital has been studying multiple myeloma, a cancer that affects the bone marrow. Their work focuses on the activity of cells in terms of their messenger RNA: this is the transcriptomic approach, or the study and analysis of DNA transcription into messenger RNA.
The combination of single-cell technology and transcriptomics is referred to as single-cell RNA-seq. “Thanks to this, it is possible to differentiate subtypes of cells in the immune system, whereas previously you could only see a group. Thus we can now look at the complex responses involved and how a tumour cell modifies its environment,” explains Minvielle. The single-cell technology can also be mixed with other experimental protocols. Vallot studies epigenetics, or the activity of a cell through regulation mechanisms of gene expression. “We apply the single-cell method to epigenetics, and only a few laboratories in the world are doing this”, she explains. The amalgamation of these two techniques is referred to as single-cell ChIP seq.


At the IGFL in Lyon4, in southeastern France, Yad Ghavi-Helm is working on embryos of the fruit fly, or Drosophila melanogaster. The aim is to use it as a model to understand the interactions between regulatory regions in the genome (enhancers) and genes, “because mutations in enhancer sequences can affect the expression of the genes they regulate and cause diseases such as cancer”, details the scientist. As in Nantes, Yad Ghavi-Helm associates single-cell RNA-seq with spatial transcriptomics, a technology that combines the high-throughput profiling of gene expression with information on the spatial localisation of cells in their original tissues or organs. “Until now, only imaging methods enabled us to study the expression of genes in this way, but this technique remained limited to a handful of genes per experiment. With spatial transcriptomics, we can analyse hundreds, if not thousands, of genes in parallel,” she adds.
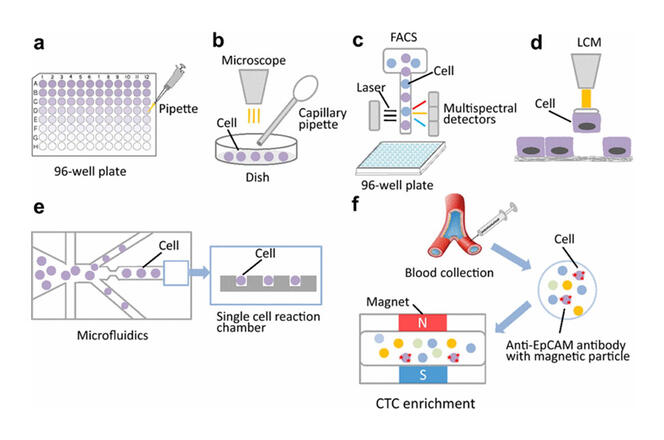


Furthermore, single-cell methods make it possible to achieve a more in-depth understanding of the role, modification and degradation of proteins in cells. Proteomics is the science that studies proteomes, or in other words all the proteins in a cell. Before the single-cell technique, obtaining access to these molecules constituted a major challenge: highly sensitive, impossible to amplify, they need to be analysed at a very small scale that renders classic methods such as mass spectrometry both imprecise and inefficient. By starting from the scale of the single cell, this technology applied to proteomics (SCP) can address questions hitherto unexplored using previous methods. Single-cell technology is advancing so rapidly that its technical limitations are on the point of being overcome, but the cost of its use remains a major obstacle for laboratories.
Turning life into equations
“With the introduction of single-cell methods, we have moved from an Excel table with 20,000 lines to calculation servers,” confirms Minvielle. The technology has given rise to a broadening of data generation: “those who produce the information can no longer interpret it alone, so biologists need support from both engineers and methodological research in statistics and artificial intelligence”, explains Franck Picard, CNRS research professor in the Laboratory of Biology and Modeling of the Cell (LBMC)5 and specialist in statistics and machine learning, which are central to the single-cell revolution. Vallot wholeheartedly agrees: “I have the impression that we are using only 5% of what this data is telling us. If we were as skilled as ChatGPT in analysing our data, that would be great. But how can we equip ourselves with the resources and machines that will derive a maximum of information from this data?” Single-cell methods imply considerable interdisciplinarity, creating bridges between biologists and those conducting research in data analysis; attracting mathematicians to biology is therefore becoming a necessity.
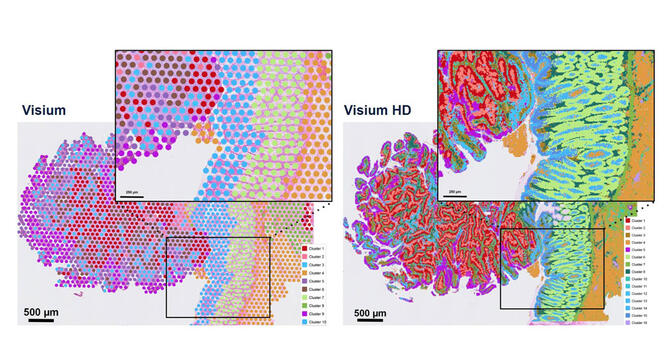

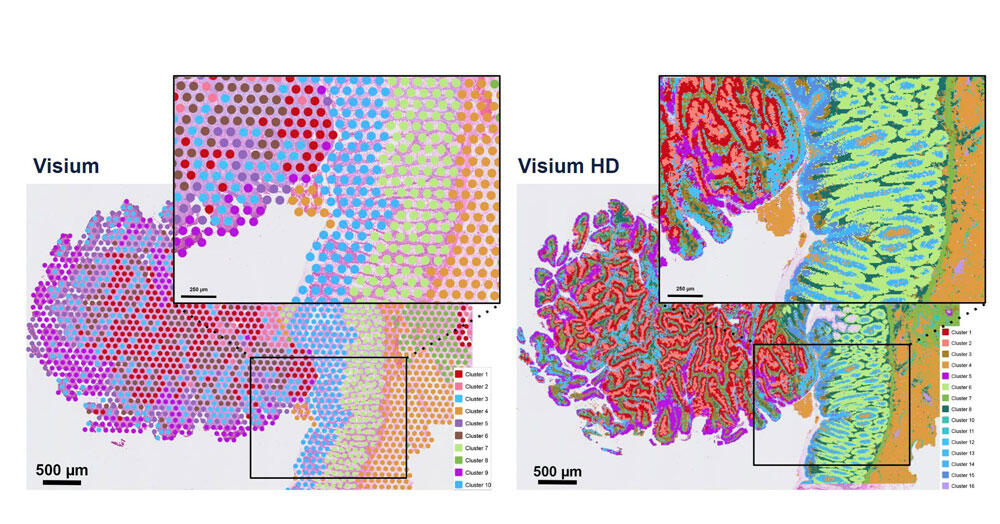
“The challenge for methodological research is to turn living organisms into equations,” summarises Minvielle. “Our advantage in France is that training in maths and machine learning is excellent,” adds Franck. But there is fierce competition in the field of artificial intelligence (AI) as students tend to be recruited by the private sector where they can earn much more than in public research. To remedy this, Picard and Vallot are banking on PhD training. “We need to value and encourage PhD projects to build these hybrid skills over the long term”, confirms Picard, with a view to developing more interdisciplinary institutes. Under this dynamic, single-cell technology has been showcased since 2023 in the digital health priority research programme and facility6, managed jointly by the Inserm and Inria. This consortium brings together experts in single-cell data analysis. Picard, who is coordinating one of the projects in this programme for the CNRS7, explains that “the aim is to develop new AI methodologies for the exploitation of single-cell data. The long-term objective is to create a link with precision medicine and, above all, to ensure the emergence of a French community specialised in this field”. The revolution is only just starting. ♦
- 1. Dynamics of Genetic Information: Fundamental Bases and Cancer (DIG-CANCER – CNRS / Institut Curie / Sorbonne Université).
- 2. Tang, F., Barbacioru, C., Wang, Y. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6, 377–382 (2009). https://www.nature.com/articles/nmeth.1315
- 3. CNRS / INSERM / Nantes Université / Université d'Angers.
- 4. CNRS / ENS Lyon.
- 5. CNRS / ENS Lyon.
- 6. https://pepr-santenum.fr/en/home/
- 7. MultiScale AI for SingleCell-Based Precision Medicine (AI4scMED) https://pepr-santenum.fr/en/2023/11/06/ai4scmed-en/