You are here
The new frontiers of the living world

How long has 'modern' biology been around?
Catherine Jessus:1 Biology as we understand it today is relatively recent. It results from the merger at the end of the nineteenth century of two major currents originating in antiquity, the naturalists and the physiologists. The naturalist movement, one of whose earliest representatives was Aristotle and then more recently scientists such as Linnaeus, Lamarck and even Darwin, never ceased to observe, describe and methodically record the living world. The beginnings of naturalism are usually situated in antiquity, and yet when you look at France’s Lascaux and Chauvet caves, you can see animal representations from prehistoric times whose precision and accuracy show that our distant ancestors were keen observers of the nature around them.
The second approach, that of physiologists with a medical background, originated with Hippocrates and Galen and was embodied in the nineteenth century by scientists like Claude Bernard. It sought to understand how the human body functioned: the role blood plays, how the heart works, and so on. These two currents came together in the early twentieth century, when naturalists started to use the physiologists' experimental methods and apply them to zoology and botany. This marked the emergence of biological science in the strict sense of the term, no longer focusing solely on observation but placing experimental methods at the heart of its intellectual approach: a hypothesis was formulated and an experimental protocol devised to address the question.
What led to the great advances made by the new science of biology during the twentieth century?
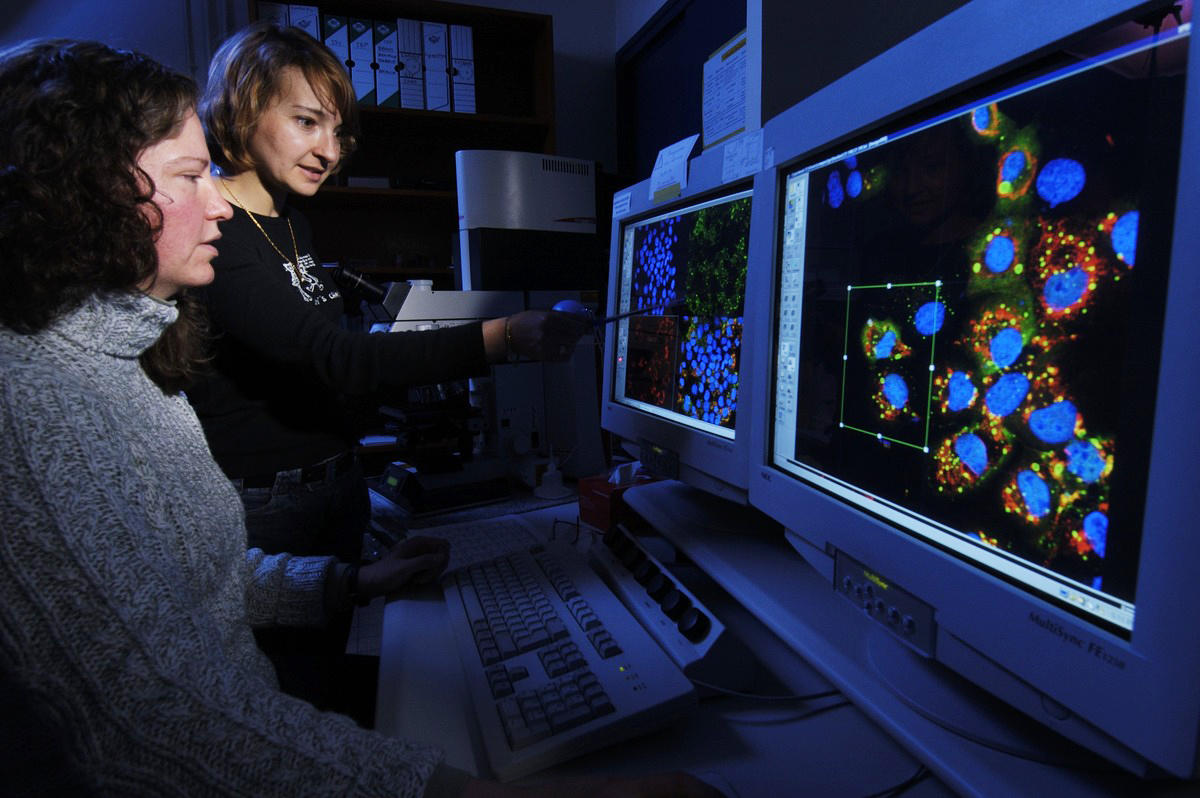
C. J.: It was essentially driven by the tremendous progress in instrumentation. The giant strides in imaging and microscopy accomplished throughout the twentieth century made it possible to see right into the interior of the cell itself. However, the biological material had to be 'sliced up' before being put under the microscope, in other words, you had to kill it before you could observe it. Today, on the other hand, biologists can use fluorescent markers to label a molecule and track it inside a living organism. The emergence of molecular biology in the 1950s brought with it fundamental knowledge about the structure and function of the genome, and laid the groundwork for its manipulation.
With the arrival of metagenomics in the past twenty years, we now have a complete picture of all the genomes contained in a sample taken from the environment, whether it's sea or river water, soil, or even the intestinal contents of a living thing. This minor revolution was made possible by the advent of big data (in other words, the processing of mass data), which is beginning to lift the veil on the endless diversity of the microbial world we know so little about. In addition to these advances, I would add the very recent progress in genome engineering brought about in particular by tools such as the CRISPR/Cas9 genetic scissors, as well as in tissue and cell engineering, which make it possible to study the functioning of a single cell. Although all these achievements are impressive, the scope of what remains to be discovered is constantly expanding.
Does this mean that the more we discover about the living world, the more we realise how little we know about it?
C. J.: Yes, I suppose you can say that. Take the example of the DNA molecule, which was discovered in the 1950s. In humans, it is two metres long, and yet it fits inside a cell nucleus a mere five micrometres across. Besides the amazing feat of cramming such a large amount of material into such a small space, it is important to understand that, of those two metres, only 6 centimetres are what's known as coding DNA, in other words, DNA that carries the genes that code for the production of proteins. All the rest is made up of non-coding DNA, which we used to call 'junk DNA', until we realised a few years ago that it is transcribed into non-coding RNA, whose importance we are only just beginning to get a glimpse of. These RNAs help plants to fight viral and bacterial infections, for example, and it is a large strand of non-coding RNA that wraps around one of the two female X chromosomes to block its expression.
What then are the major challenges for the biologists of the twenty-first century?
C. J.: I would mention the exploration of the microbial world, including organisms living in extreme environments and novel, unexpected forms of life; the study of the drivers of evolution, going beyond mere point genetic mutations; the functioning of genomes, which we've just talked about; the study of interactions between living beings, and between living beings and the environment – looking at organisms in isolation makes no sense, knowing that all life forms, from bacteria to humans and plants, are integrated into delicately-balanced, complex systems; and finally, cognitive functions and the brain, as well as the immune system, a subject too vast to be covered in this interview. And this list is far from exhaustive!
Let's start with the definition of what constitutes life. You told us that it has undergone changes in recent years. What led to this reappraisal?
C. J.: Until the early twenty-first century, it was thought that life couldn't develop in extreme conditions: above certain thresholds of temperature, pressure, salinity and so on, or in the absence of light or oxygen. The discovery of living things in such environments – in boiling lakes or geysers, in the darkness of the deep sea, and in acidic media such as the volcanic site of Dallol in Ethiopia – again raises the question of what life is.
These extremophile organisms, which are mostly microorganisms, have metabolisms, cell membranes and other previously unsuspected characteristics. They should also help to shed light on the origins of life on Earth, 3.8 billion years ago, and on what we should be looking for on other planets. In the meantime, studying living beings in these extreme habitats presents real technical challenges for researchers, currently unable to culture most of these life forms in the laboratory.
Let's turn to the world of microbes, which, we are told, is a genuine terra incognita...
C. J.: The microbial world continues to elude us, even in environments that are more familiar and closer to home than the ones we've just been talking about. As a result of the Tara Oceans expedition organised by the biologist Eric Karsenti, we have discovered in particular that the oceans are inhabited by a plethora of microorganisms: it is estimated that 60% of the bacteria present on our planet live in the sea, and yet only 5% have been described! A huge amount of work identifying and cataloguing them remains to be done, comparable to the achievements of naturalists in the past. We also need to take a fresh look at microorganisms. We tend to see them solely as pathogens (even though only a tiny minority of bacteria and viruses are harmful to humans), whereas on the contrary, they are key to maintaining the balance of the planet, a role we're just beginning to glimpse.
It is hard to believe that it is only a few decades since we discovered the domain of the Archaea, the third branch of life, along with Bacteria and Eukaryotes. Although found everywhere in extreme environments, archaea are in fact encountered in all ecosystems. However, we know very little about them and are only starting to explore their genomes, which are very complex and contain many genes with unknown functions… And then there are the giant viruses discovered at the turn of the twenty-first century!
What do we know about these giant viruses?
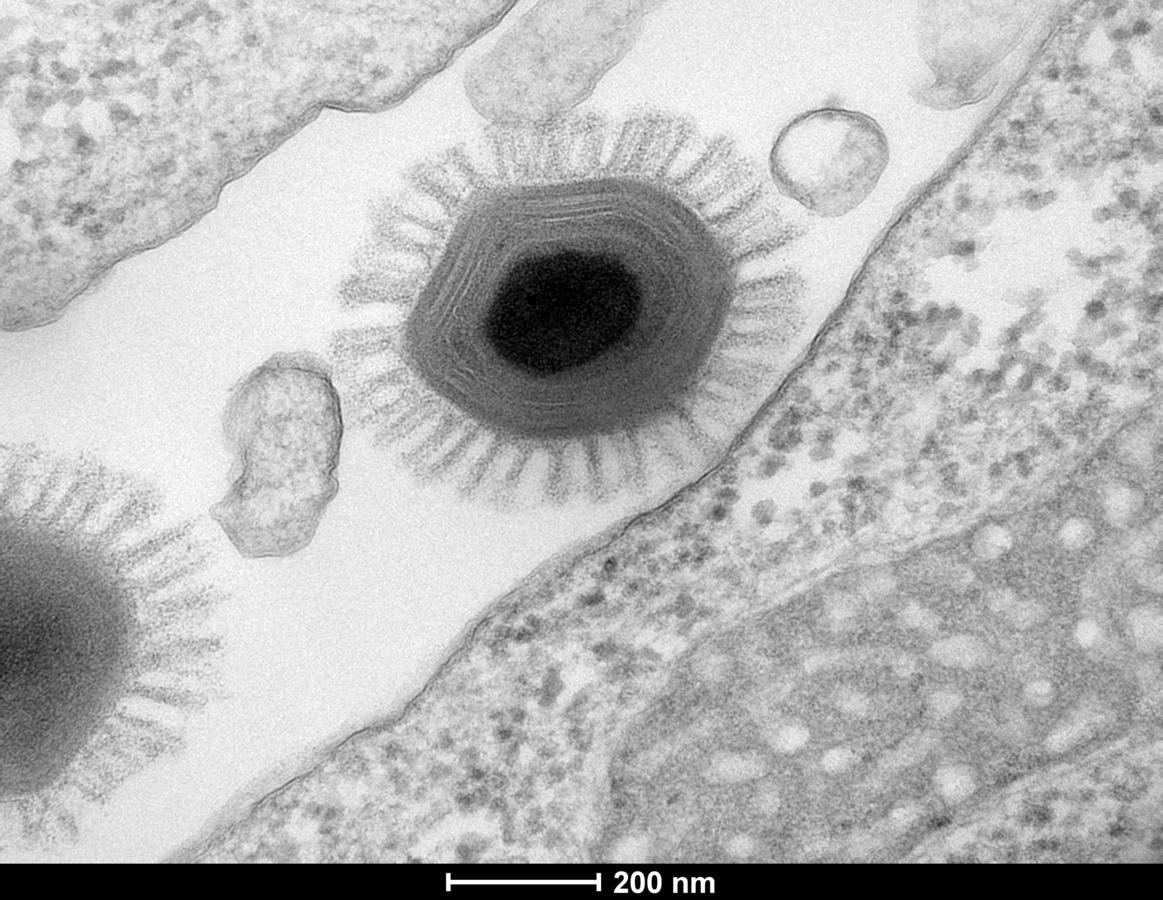
C. J.: The term 'virus' is in fact a misnomer. Like normal pathogens, they are parasites that cannot reproduce outside a host cell. But unlike ordinary viruses, they have huge, complex genomes, made up of thousands of genes; they are as large as bacteria; and they have a very unusual envelope. Where have they come from? How should we characterise them, since they are neither viruses nor cells? The subject has given rise to intense discussion. In any case, they are found all over the planet, living in the amoebae present in water everywhere, including tap water. As far as we know, they are not pathogenic, but then we're a very long way from knowing them all! And then there are the megaviruses imprisoned in the permafrost over the last 30 000 years, which are gradually being released as global warming thaws the frozen ground, and with which modern humans have not coevolved.
Why do you think we should focus on the drivers of evolution in the coming decades?
C. J.: It was long thought that new species were created through point mutations in genes. But in fact, this is just one of many ways in which new organisms can come about. The main source of innovation lies in the modification of large sections of genomes, which happens, for example, via the exchange and fusion of genomes between species. For instance, we now know that the first eukaryotic cell emerged through the combination of an archaeon and a bacterium. The bacterium that had entered the archaeon gradually lost its autonomy, allowing the formation of the membrane compartments, especially the mitochondria, that characterise eukaryotes. The same thing applies to the emergence of plants: a eukaryote entered into symbiosis with a bacterium that was able to use light to produce energy, giving rise to the first microalgae. And it was this same mechanism that explains why mammals have a placenta that enables the embryo to develop within the mother's body.
You mean mammals are also the result of gene exchange?
C. J.: Exactly. The placenta developed thanks to the transfer of genes from a retrovirus to an animal that laid eggs. This particular retrovirus was able to produce proteins causing cell fusions, and it was these proteins that gave rise to the placenta. Without this little retrovirus, we'd still be laying eggs! Examples like these demonstrate the importance of the exchange of genetic material between living beings, something that was long neglected.
Interactions are the next great challenge for biology in the years to come.
C. J.: Relationships between organisms are essential for life. All living things are either contained within a larger organism or contain smaller ones – none of them exist in isolation. This is well illustrated by the intestinal microbiota, which is made up of archaea, bacteria, viruses and yeasts. It can weigh up to 2 kg and functions as one of our organs – which, by the way raises real questions about what is 'self' and 'non-self'. The microbiota is essential for the functioning of our digestive system, and without it, we wouldn't be able break down all the food we ingest. It is also necessary for the maturation of our immune system, and secretes key molecules that affect the way our organs work, including the brain!
Any imbalance in this system can cause problems. For instance, a bacterium in the microbiota that starts to proliferate, taking advantage of the decline of other microbial species, can very quickly become malignant. In fact, we are only just beginning to discover the complex nature of the living world, in which everything is a question of proportion and balance, whether on the scale of an individual, a population or an entire ecosystem. It is this complexity that we need to address today. Investigating cells, tissues, organs or individual organisms in isolation is a thing of the past: today biologists need to study entire networks made up of myriads of players.
The last example I'll mention, and undoubtedly the most striking, is that of the brain, an organ of staggering complexity due to the number of cells involved, and where there's nothing but networks and interactions. Thanks to imaging data, we're beginning to have a pretty clear map of the brain and understand not only which region interacts with which, but also how electrical information flows at the nanometre scale of the synapse. But the real challenge for biologists remains the cognitive brain: the memory, recall, emotions, decision-making, and all the chemistry that drives these processes. In this domain, too, there is still an awful lot to discover.
- 1. CNRS senior researcher at the Developmental Biology Laboratory (CNRS / Sorbonne Université), where she heads the Oocyte Biology team. She directed the CNRS's Institute of Biological Sciences from 2013 to 2019.