You are here
Epigenetics Rules the Genome

During the past ten years or so, epigenetics has been riding high. This branch of biology focuses on the molecular mechanisms that can regulate the expression of genes without modifying the nucleotide sequence from which they are derived. Epigenetics does not modify the partitioning of the genetic code but the way it is read, and therefore acts as a sort of conductor of the genome. Surprising as it may be, identification of the very first factors for epigenetic regulation—proteins in the Polycomb group—preceded that of the double helix structure of DNA. Indeed, their demonstration by the American biologist Pamela Lewis dates from 1947 (while James Watson and Francis Crick posited the double helix of DNA in 1953: editor’s note). It was by examining the dominant mutations in Drosophila that she discovered—almost by chance—the genes coding for these protein complexes. Based on the pioneering work by his wife, the celebrated geneticist Edward Lewis1 then showed in the late 1970s that Polycomb proteins restrict the expression of genes involved in the organizational plan of living beings. During subsequent years, other studies in Drosophila revealed the existence of Trithorax proteins, whose mode of action is opposite to that of Polycomb proteins.2
Biochemistry to the rescue of genetics
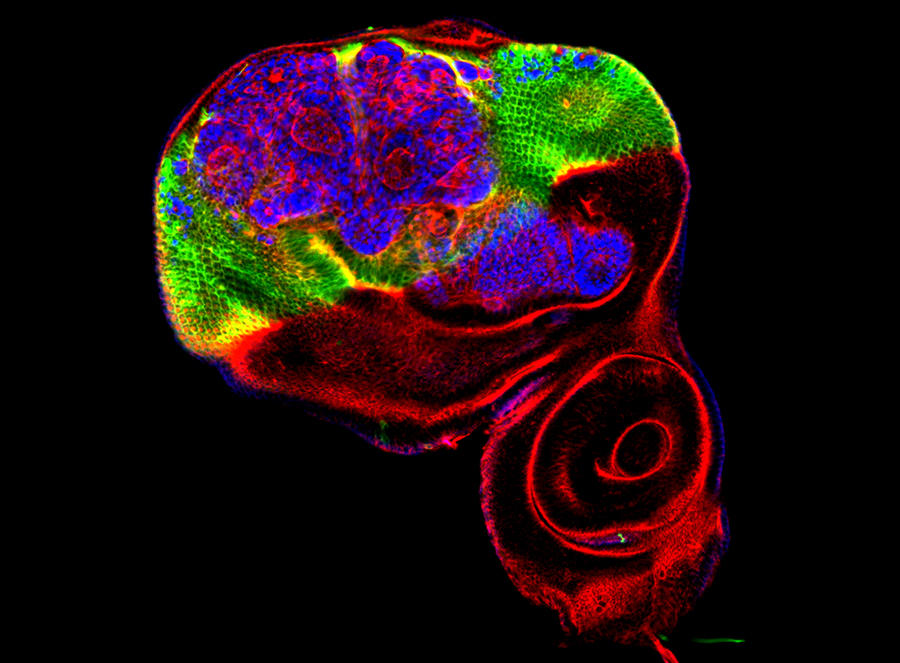
Despite promising early findings, study of these epigenetic regulatory factors was then somewhat neglected for several decades. It was only in the early 2000s that their mode of functioning was precisely identified. Thanks to advances in biochemistry, it was then discovered that the structure and function of Polycomb and Trithorax proteins had been conserved during evolution, from plants to fungi to animals. “By becoming involved in the field of genetics, biochemistry made it possible to characterize these protein complexes at the molecular level and thus understand that they intervene in epigenetic modifications of tails of histones, the proteins around which DNA winds itself to be compacted within the cell nucleus,” explains Andrew Saurin, a CNRS researcher at the Developmental Biology Institute of Marseille (IBDM)3 in southeastern France where he leads his own research team. These regulatory mechanisms, which are similar to simple chemical modifications to the chromatinFermerthe basic structural unit of chromosomes, made up of the DNA molecule with which are associated RNA and proteins, were first of all identified at the level of homeotic genes. Sometimes referred to as architect genes, they ensure the correct arrangement of different organs throughout the development of most animals. In this context, the Polycomb proteins are specifically responsible for preventing the expression of homeotic genes, while Trithorax proteins activate them in certain types of cell when this proves necessary to ensure growth of the individual.
A form of anti-tumor regulation
“The principal function of these two molecular complexes is to maintain the memory of the regulatory status of genes through cell divisions from the embryonic stage up to the adult organism,” details Giacomo Cavalli, CNRS senior researcher at the Institute of Human Genetics (IDH)4 in Montpellier (southern France). In his laboratory, this molecular biologist has been trying for several years to clarify the subtle modes of functioning of epigenetic regulatory factors. Building on an innovative very high-throughput sequencing method, Cavalli and his colleagues have succeeded in mapping in considerable detail the three-dimensional organization of all of the chromosomes in Drosophila. They discovered that this 3D structure can be transmitted from generation to generation, thus contributing to maintaining epigenetic regulation of the genome.5 More recently, and in the same model organism, the team managed to demonstrate the anti-tumor function of a protein complex in the Polycomb family.6 “By binding specifically to hundreds of genes involved in the formation of tumors, such as those that control cell proliferation, this combination of proteins manages to regulate both their temporal and spatial action,” explains the scientist. “We were also able to observe a form of anti-tumor regulation that was similar in human cell lines in culture.”
An essential link between different mechanisms
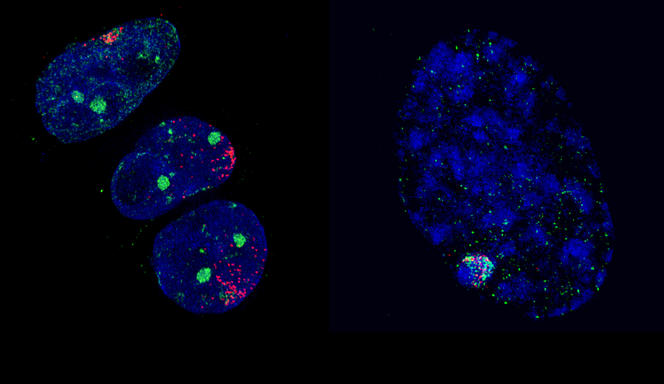
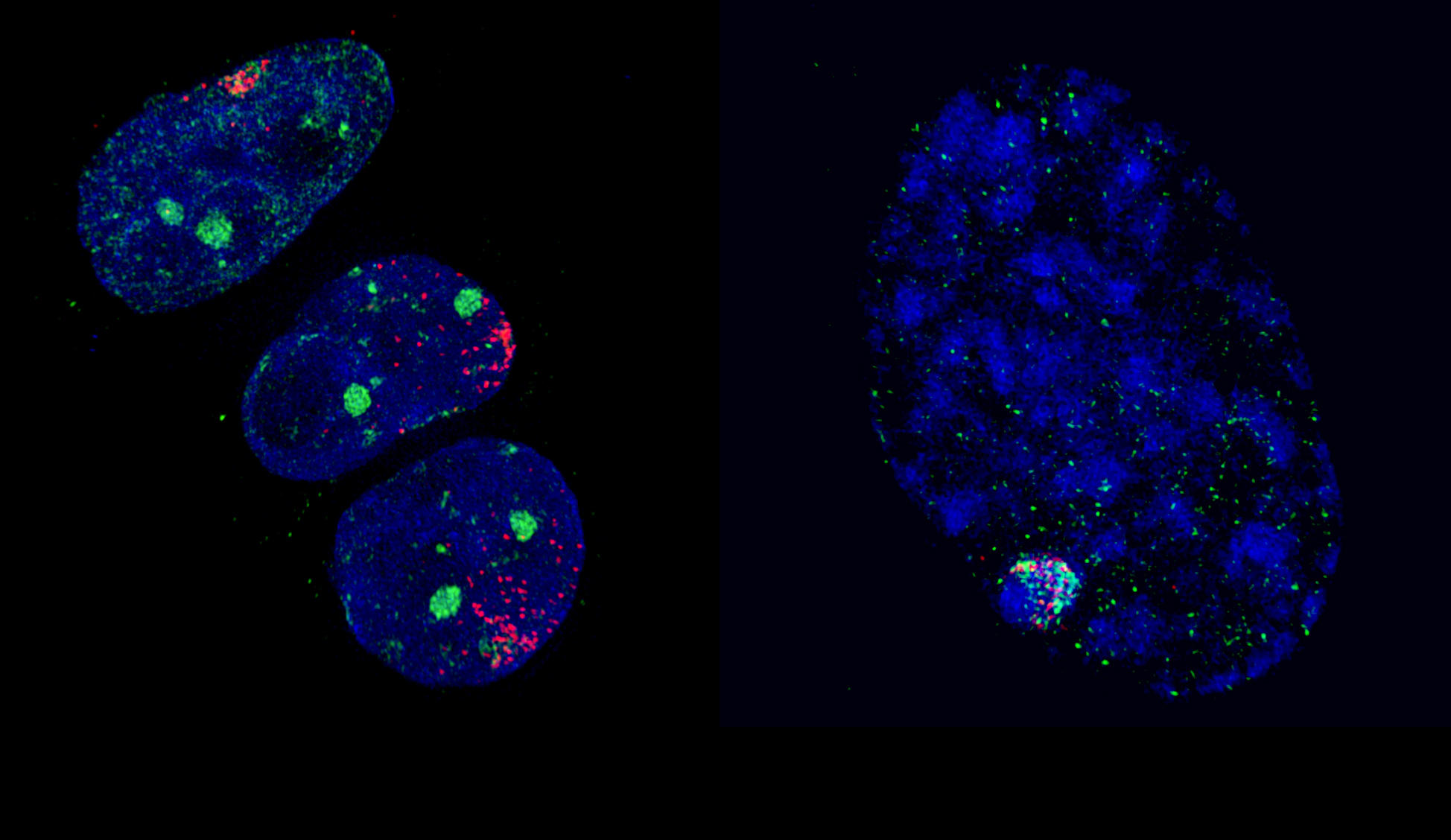
Along with these findings, scientific studies demonstrating that Polycomb proteins play a key role beyond simple developmental processes have proliferated in recent years: stem cell plasticity and renewal, determination of the fate of cells, inactivation of the X chromosome, etc. This latter regulatory mechanism was the favourite research topic of Edith Heard, Professor at the Collège de France where she holds the Chair of Epigenetics and Cellular Memory. In this field, work by the geneticist has contributed to elucidating how, in female mammals, one of the copies of the X chromosome from either the father or mother is inactivated in a random manner in each cell during embryogenesisFermerin animals, embryogenesis corresponds to the developmental phase of the organism from fertilization to birth. Inactivation of the X chromosome, which represents a reliable model for the study of epigenetic regulatory mechanisms, has notably made it possible to establish new rules for the interactions between Polycomb proteins and chromatin. “In some types of cancer, it appears that the close association between the inactive X chromosome and Polycomb regulatory factors is disturbed, so that the latter are then dispersed throughout the cell,” explains Heard, who also heads the Génétique et Biologie du Développement7 Unit at the Institut Curie. In this laboratory, the scientist and her team are pursuing their investigations into inactivation of the X chromosome. Like other research groups working in close collaboration with clinicians at the Institut Curie, they hope to determine the principal types of epigenetic deregulations that occur in the context of cancer, in order to establish a link with their associated genetic mutations.
Two-thirds of genes are affected
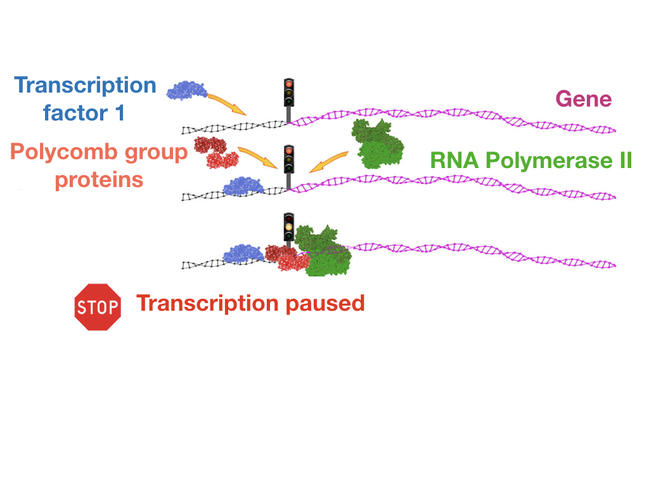
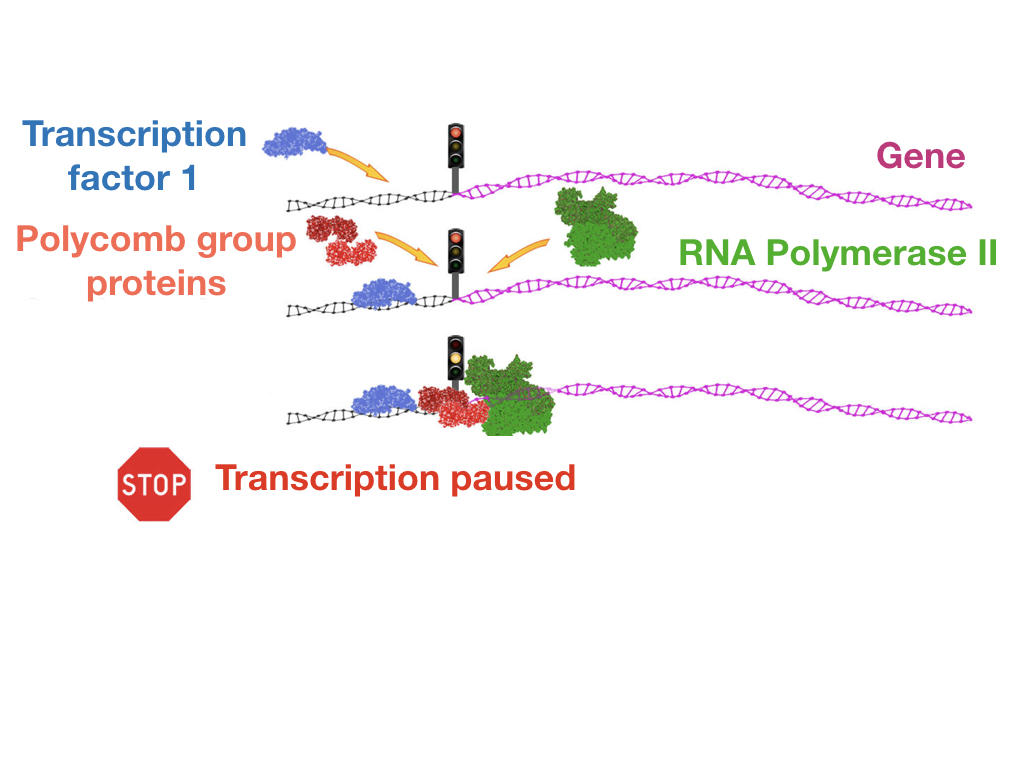
In a completely different area, Polycomb proteins now appear to be essential levers for controlling the transcription of a very large number of genes. These original types of regulation, whose existence was still unknown some 15 years ago, are now the subject of regular publications, such as those generated by the work of Saurin’s team at the IBDM. In a study published last year,8 these scientists showed that certain protein units in the Polycomb group were able to prevent the action of polymeraseFermerenzymatic complex involved in the transcription of DNA into RNA by binding to promoting sequences, the DNA regions located near the genes that are essential for the transcription of DNA into RNA. “As far as scientists are aware, it is estimated that the type of gene expression regulation involving Polycomb concerns nearly two thirds of the 20,000 genes in our genome,” underlines Saurin. These notably include metabolic genes whose deregulated expression is known to cause the onset of cancer. “Although the stable and heritable repression of homeotic genes by Polycomb is essential (because it determines the organizational plan of individuals throughout their lives), it nonetheless remains a specific case,” he explains. “It is therefore in the more general context of transcription regulation—where the role of Polycomb has been little explored—that the principal modes of action of these proteins probably lie, notably in the area of carcinogenesis.”
Therapeutic avenues
Unlike the genetic mutations that directly affect our genome, epigenetic marks result in simple chemical modifications of DNA or its associated proteins within the chromatin. These marks do not change the nucleotide sequence, so it should be possible to modify them without preventing the gene concerned from expressing itself correctly, which thus raises new hopes for the treatment of certain diseases. Heard is measured in her opinion: “although it has been possible to highlight numerous epigenetic aberrations in different types of cancer, their respective roles are still poorly understood,” she says. However, scientists reckon that these abnormalities may cause the inactivation of tumor suppressor genes or, inversely, the activation of oncogenes, whose over-expression favours the development of cancer. “In the case of Polycomb, the possibility to neutralize the active site of the protein responsible for its negative effects opens the way to new types of treatments,” asserts Cavalli. “Molecules tested in clinical trials have indeed produced encouraging results for the treatment of circulating tumors such as lymphomas, as well as certain solid tumors.” But while a strategy of this type appears to have a beneficial effect on the development of several types of cancer, it may also prove totally ineffective, as was recently demonstrated in a study carried out by scientists at the Institut Curie.9 Although the prospect of treating a broad range of pathologies using molecules that target epigenetic mechanisms seems attractive at first sight, it is wishful thinking as the mode of action of such treatments, and their effects on future patients, are still little known.
- 1. Edward Lewis was joint laureate of the Nobel Prize in Physiology or Medicine in 1995 for his work on the genetic control of early embryonic development.
- 2. B. Schuettengruber et al.,“Genome Regulation by Polycomb and Trithoilrax: 70 Years and Counting”, Cell, 2017. Doi: 10.1016/j.cell.2017.08.002
- 3. CNRS/Aix Marseille Université.
- 4. CNRS/Université de Montpellier.
- 5. F. Ciabrelli et al.,“Stable Polycomb-dependent transgenerational inheritance of chromatin states in Drosophila,” Nature Genetics, 2017. Doi :10.1038/ng.3848.
- 6. V. Loubière et al., “Massive Coordinate redeployment of PRC1 proteins suppresses tumor formation during Drosophila development,” Nature Genetics, 2016. Doi:10.1038/ng.367.
- 7. CNRS/Institut Curie/Inserm /Sorbonne Université.
- 8. A. Zouaz et al., “The Hox proteins Ubx and AbdA collaborate with the transcription pausing factor M1BP to regulate gene transcription”, EMBO Journal, 2017. Doi: 10.15252/embj.201695751.
- 9. M. Schoumacher et al., “Uveal melanoma cells are resistant to EZH2 inhibition regardless of BAP1 status”, Nature Medicine, 2016. Doi: 10.1038/nm.4098
Explore more
Author
After first studying biology, Grégory Fléchet graduated with a master of science journalism. His areas of interest include ecology, the environment and health. From Saint-Etienne, he moved to Paris in 2007, where he now works as a freelance journalist.