You are here
The dawn of digital oncology

How can the detection of certain tumours be improved? Can their evolution or response to treatment be predicted? What is the mechanism that causes the proliferation of cancer cells? These are some of the questions that the researchers at the Michel Laudet digital oncology laboratory is trying to answer. Created officially in April 2022, this unit results from collaborative efforts over several years between the Irit computer science institute in Toulouse1 and the Toulouse Cancer Research Centre (CRCT)2. “Biomedical and clinical data has become so abundant that it can no longer be processed by the human brain. Only artificial intelligence (AI), combined with the expertise of physicians, is now able to sort, classify and above all exploit this mass of information,” explains Jean-Marc Alliot, scientific director for Big Data and artificial intelligence at Toulouse University Hospital (CHU).
This assessment is shared by Professor Pierre Brousset, who is a member of the Académie de Médecine, Director of the Toulouse Cancer Laboratory of Excellence (Labex Toucan), and head of the pathological anatomy and cytology department at the IUCT-Oncopole3. He has no doubts that “oncology is now entirely dependent on computer scientists, and not the other way around! Biostatistics, bioinformatics through the high-throughput sequencing of tumour genomes, and artificial intelligence are so many digital tools that are constantly enriching our knowledge of cancer and renewing our therapeutic strategies”.
Tools to aid diagnosis
Based on the fourth floor of the imposing CRCT building, the dozen or so Irit scientists specialise in the design of bio-inspired algorithms (reproducing life), machine learning software and signal processing and analysis (images and sound). “Most of these scientists have already completed projects in the medical research field,” explains Sylvain Cussat-Blanc, an Irit scientist and member of the ANITI4. “The advantage here is that we can work in constant interface with researchers, which drives and accelerates our projects.” Since the Michel Laudet digital oncology lab was set up, some fifteen projects have been initiated or are underway. All are jointly led by a computer scientist and a physician or medical researcher. “As with any multidisciplinary work, the principal challenge consists in finding a common language to clearly define the question that the researcher wants us to answer, and determine the possibilities and constraints of artificial intelligence,” Alliot adds.
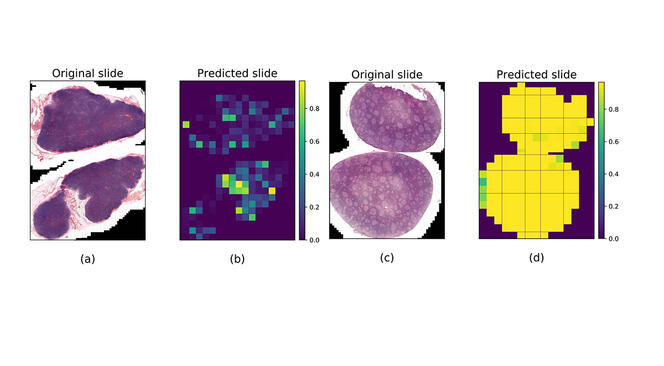

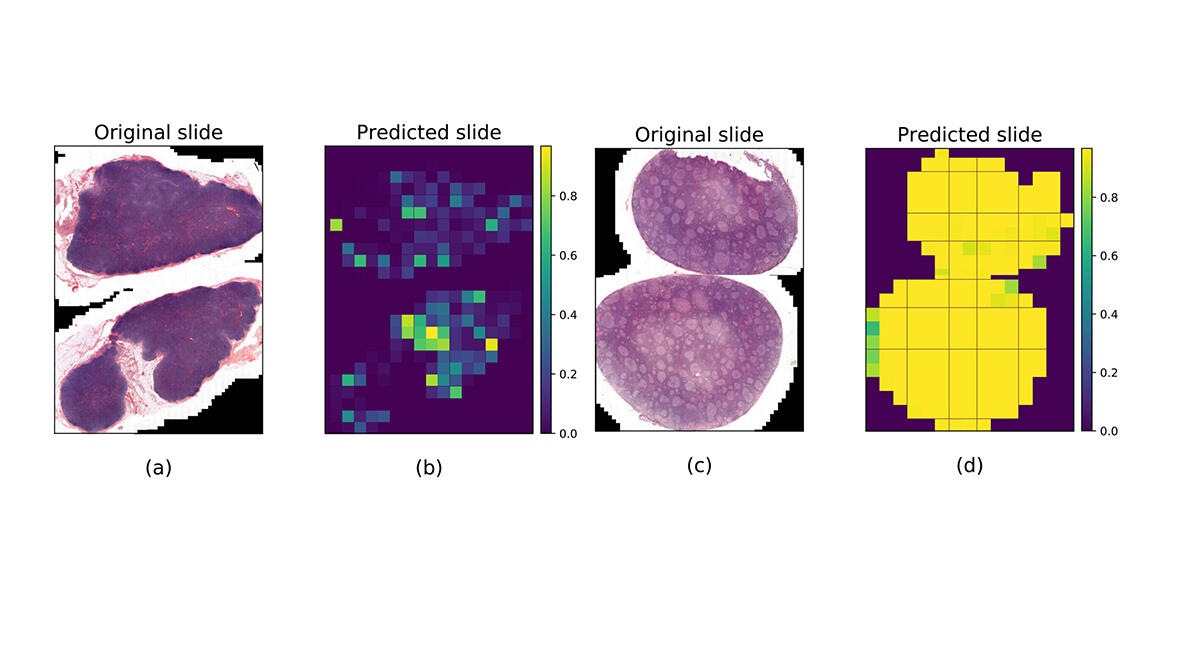
Machine learning algorithms are the most requested by research physicians and particularly by pathological anatomists, for whom the issue of diagnostic support is crucial. “The management of cancer is based to a large extent on the microscopic examination of biological tissues by a pathological anatomist,” explains Brousset, who specialises in lymphomas. “Some lesions are complex and difficult to characterise, which can lead to serious diagnostic errors.”
In 2020, this scientist contributed to creating a machine learning algorithm5 to differentiate follicular lymphoma – a cancer of lymphocytes in the lymph nodes – and follicular hyperplasia (a benign disease), since these two lesions often display similar characteristics. To train the software to recognise them, 378 whole slide images from Toulouse University Hospital were used; half of them were of lymph node tissues infiltrated by follicular lymphoma, while the other half were affected by follicular hyperplasia. The result was that in 99% of cases, the software achieved a precise diagnosis of the lesions, “which surpassed the performance of the pathologist using his microscope alone”, Brousset admits.
The trained algorithm was then tested on images from Dijon and Montpellier University Hospitals (in eastern and southern France, respectively), but the success rate only reached 65%. “This difference came as no surprise,” notes Jean-Marc Alliot. “For as long as medical data from hospitals is not produced for the specific use of algorithms, applying uniform methods and formats, there will be bias that alters the performance of AI”. The application may nevertheless be tested experimentally at Toulouse University Hospital in the coming months.
Guiding therapeutic decision-making
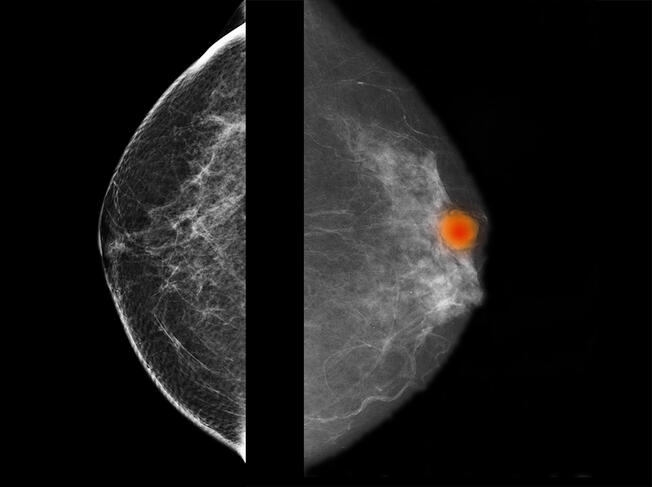
Several similar diagnostic support projects are currently ongoing. They concern the identification and recognition of potentially cancerous oral lesions, and also certain breast tumours that are difficult to characterise. Working with Camille Franchet, a pathological anatomist at IUCT-Oncopole, the team at Michel Laudet is participating in the APRIORICS deep learning project reinforced by immunohistochemistry to requalify images of breast cancers6, and aims to use artificial intelligence to describe the tumours as precisely as possible based on a large number of parameters. The aim is to develop predictive models that will optimise diagnosis.
In 2021, an algorithm was designed with the haematologist Sarah Bertoli to predict the survival of patients with acute myeloid leukaemia. In this cancer, which develops in the bone marrow, the question was to try and determine the survival prognosis of patients at a given time with the two most widely-used treatments. To train the software, clinical, genomic and therapeutic data on 5,000 patients followed for 20 years at Bordeaux (southwestern France) and Toulouse University Hospitals were homogenised and processed. The results7 showed that in nearly 70% of cases, the survival prognosis proved correct. In 90% of cases, based on medical data at the time of diagnosis, the learning algorithm was even able to choose the same treatment as that recommended by the expert haematologist.
A new study has just been launched with Professor Marlène Pasquet (haematologist and immunologist at Toulouse University Hospital and researcher at the CRCT), this time to predict the course of a rare form of acute myeloid leukaemia in young patients and to define the best therapeutic option depending on their molecular profile. “These algorithms are not designed to replace a physician, but to support the personalised management of patients in the longer term,” Brousset stresses.
Describing biological processes and finding new therapeutic options
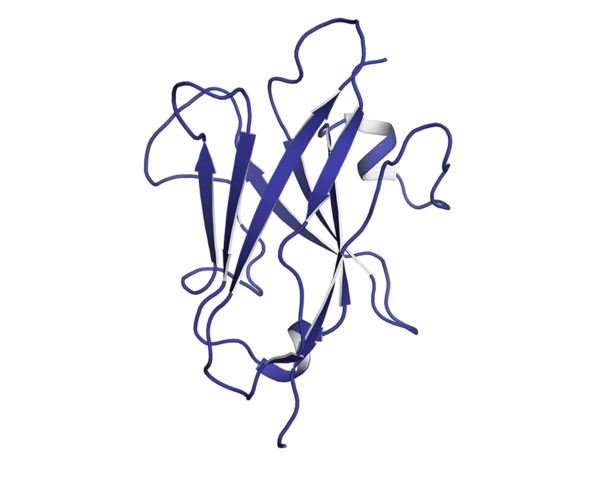
“Artificial intelligence is limited to deep learning,” insists David Simoncini, an Irit computer scientist who specialises in the modelling of new therapeutic proteins. “There is a whole branch of AI based on logic which concerns the simulation and optimisation models used when a scientist wants to clarify complex biological phenomena. They are particularly powerful in terms of throwing new light on certain interactions, and testing or suggesting hypotheses.”
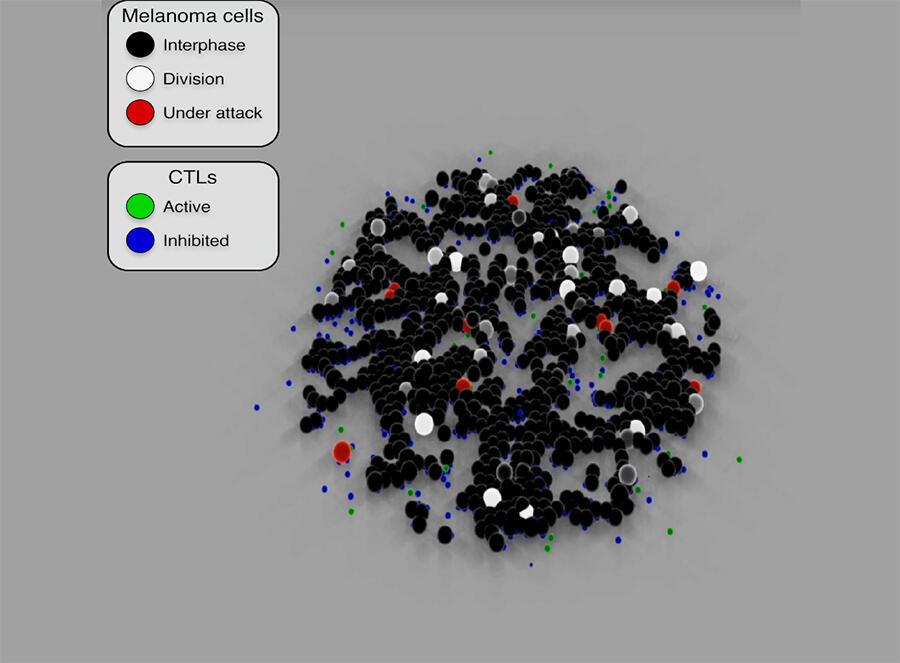
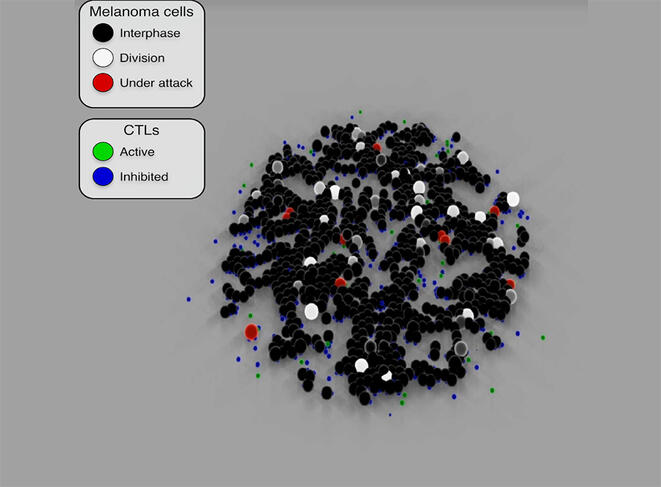
These models are able to simulate the proliferation of cancer cells and predict their evolution or their response to cancer treatment. By reproducing the attack mechanisms of cytotoxic T-cells8 (used in immunotherapy) against cancer cells, Cussat-Blanc has been able to model the action of a treatment at different doses in order to optimise its efficacy on tumour cells. He has also demonstrated9 the effect of Nocodazole (a cancer drug) on cancerous colon cells and revealed certain effects on the cellular life cycle that were previously unknown.
The scientist has been working for several years with Pierre Cordelier, deputy director of the CRCT and a specialist in oncolytic viruses in the context of pancreatic cancer (one of the most deadly). He explains: “Not only do oncolytic viruses target cancer cells while sparing healthy cells, but they are also able to stimulate the immune system, which means that by combining viral therapy with immunotherapy, it might be possible to achieve a stronger oncological effect.”
Having designed an algorithm that reproduces the infection of cancer cells by an oncolytic virus, the two scientists integrated cytotoxic T-cells in the model and since then have been conducting multiple simulations based on biological data. These constant cross-disciplinary exchanges are contributing to the emergence of new working hypotheses and thus to a clearer understanding of certain biological mechanisms. “It offers a way for us to redefine how we treat patients. In this respect, the modelling of living organisms provides a new source of fundamental discoveries in oncology and will enable the optimisation of drugs and biotherapies in the future.”
More explainable algorithms
While it is often impossible to understand the precise details underlying the decisions of learning algorithms and thus the source of the result, simulation models have a major advantage: they are reliable and explicable. All the data obtained during the different stages of modelling is indeed the subject of in vitro verifications in the laboratory in order to validate the hypotheses. “Our aim is to develop algorithms that are as transparent, explainable and traceable as possible, so that practitioners can have access to AI tools whose results enable them to explain to a patient what has led them to make a particular diagnosis or decide on a treatment option,” underlines Cussat-Blanc.
To counteract the “black box” effect of deep learning algorithms, he and his colleague Simoncini have designed a new machine learning method that can function with little data and is transparent regarding its reasoning. The software is currently under test by the team and a patent has already been filed. ♦
- 1. CNRS / Toulouse INP / Université Toulouse III - Paul Sabatier.
- 2. CNRS / INSERM / Université Toulouse III - Paul Sabatier.
- 3. The Institut Universitaire du Cancer de Toulouse Oncopole brings together all oncology departments at Toulouse University Hospital and Institut Claudius Regaud.
- 4. Artificial and Natural Intelligence Toulouse Institute.
- 5. “Accurate diagnosis of lymphoma on whole-slide histopathology images using deep learning”, Charlotte Syrykh et al, npj/Digital Medicine, 1st May 2020. https://doi.org/10.1038/s41746-020-0272-0
- 6. https://www.health-data-hub.fr/projets/apprentissage-profond-renforce-pa... (in French - link is external)
- 7. https://www.sciencedirect.com/science/article/pii/S0006497121053179
- 8. “Sequential adjustment of cytotoxic T lymphocyte densities improves efficacy in controlling tumor growth” (“Modélisation des interactions entre lymphocytes T et cellules tumorales et optimisation des injections”), Roxana Khazen et al., Scientific Reports, 23 August 2019. https://www.nature.com/articles/s41598-019-48711-2
- 9. “A checkpoint-oriented cell cycle simulation model” (“Modélisation du cycle cellulaire et de l’impact d’une molécule thérapeutique sur son déroulement”), David Bernard et al., Cell Cycle, 4 April 2019. https://www.tandfonline.com/doi/full/10.1080/15384101.2019.1591125
Explore more
Author
Carina Louart is a journalist and published author specialized in sustainable development, social issues and life sciences. She has authored several books including La Franc-maçonnerie au féminin, (Belfond publisher) Filles et garçons, la parité à petits pas ; La Planète en partage à petits pas ; C’est mathématique ! (publisher: Actes Sud Junior), which...