You are here
Artificial genomes offer a promising lead
Your research field lies at the crossroads between computer science and genetics. What main issues are you investigating?
Flora Jay:1 To be precise, I would say that my research field lies between computer science and genetics, but also mathematics and statistics. Based on genetic data, I try to reconstruct the demographic history of populations. For example, this concerns how species or people separated and then mixed again, or how certain factors or events have led to the selection of certain versions of genes. My work includes ecological, historical and medical aspects.
In 2021 you published a study which caused a stir in your scientific community: it focused on the creation of realistic portions of genomes using artificial neural networks. Could you tell us first of all what an artificial neural network actually is?
F. J.: A neural network is a statistical model that helps to understand and approach complex functions. By training it on a dataset, it can learn to carry out difficult tasks.
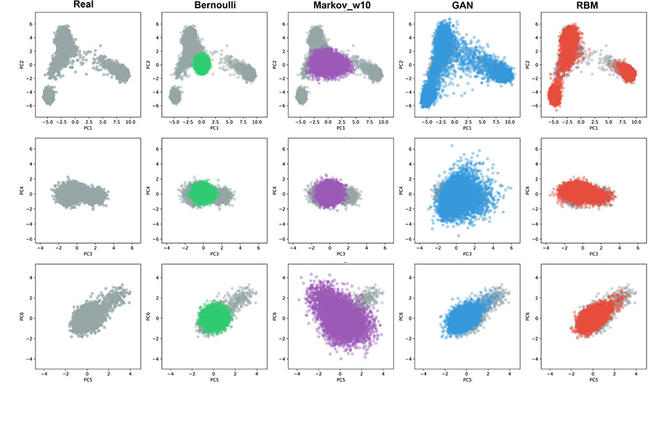
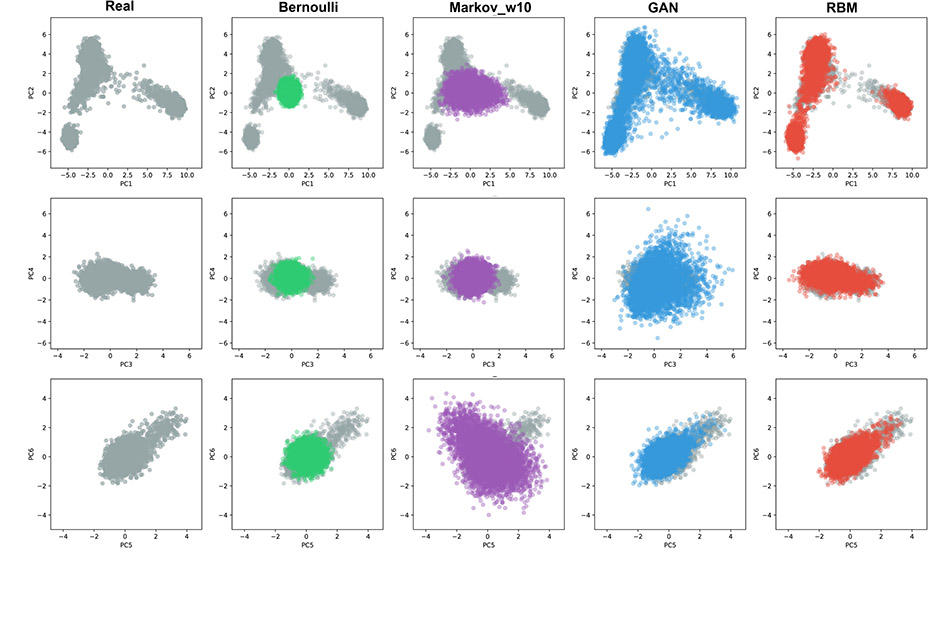
Suppose for example you want to train the network to distinguish between images of a cat and a dog. At the start, it chooses at random and is wrong half of the time. But as it continues to make mistakes and then correct itself, it finally learns to make the distinction. With Burak Yelmen, the lead author of the paper, we wanted to find out whether these machine learning methods could help us to imitate genetic data. We managed to train a neural network on a large genome databank, the Estonian Biobank, and once it had been trained, we were able to create realistic portions of genomes. These portions retained the characteristic traits of the original genomes, but in fact they did not belong to any real individual.
So you obtained a sort of photofit of the genomes in the database?
F. J.: Not quite. A photofit seeks to imitate the face of an individual. In this case, we were not imitating a particular genome. We obtained realistic sequences that were impossible to distinguish from a real genome but belonged to no-one. It was rather like computer-generated, synthetic faces that do not really exist.
What are the possible applications of these artificial genomes?
F. J.: Many genetic databanks – particularly those in private hands – do not allow scientists to have access to their data for financial reasons, and sometimes out of confidentiality concerns.Indeed, when you have numerous genomes and the population they come from is not very large, it may eventually be possible to identify who they belong to, based on family links and information such as age or characteristic traits. This raises ethical problems, because these genomes can be used to trace back – more or less reliably – to confidential data including genetic ancestry, parentage or an increased risk of being affected by a disease. Considering that publicly-available intelligence has enabled numerous discoveries in population genetics, it would be a good thing if scientists were also given access to privately-held data. Our method may offer a solution to this dilemma. Machine learning could make it possible to construct artificial genomes that would provide important information on a population under study but would not belong to any real individual. These artificial genomes could then serve biomedical or population genetics research, or indeed that in other fields.
How could that be?
F. J.: Let’s take the example of a scientist investigating which part of the Neanderthal genome is still present in a modern population. Should access be denied, for reasons of confidentiality, to real-life genetic data on this population, our artificial genomes could be used as a proxy. Created using genetic information on this particular group, they would retain these characteristics and therefore traces of this ancient DNA.
Do these methods have other applications?
F. J.: It is still highly prospective, but the training of neural networks could make it possible to identify atypical regions where selection pressures may have been exerted. For example, the mutation that enables the digestion of lactose may thus be revealed, or areas linked to resistance against pathogens. Once identified, scientists could then focus in more detail on these regions.
Could you describe your career, and what led you to work in these fields?
F. J.: I trained at an engineering school, specialising in applied mathematics and computer science, and then completed a Master’s degree in computer science and mathematics applied to biology. After a PhD in modelling and statistics for population biology, I completed a postdoctoral attachment at Berkeley (California, US) in 2012, where I worked on the DNA of two women, one Neanderthal and one Denisovan. We were trying to understand how, when, and to what degree the Neanderthals, Denisovans and Homo sapiens had mixed. I then returned to France and worked on the reconstruction of demographic history and the development of statistical methods to summarise genomes without losing too much data. I started to wonder whether it was possible to build neural architectures adapted to extracting information from genomes in a more automated manner. That is why I joined the LISN interdisciplinary digital science laboratory at the end of 2015.
What’s the driving force in your work? Is it issues related to genetics and evolutionary history, or the development of new analytical methods?
F. J.: It is difficult to answer that question. I like collaborating because when you work with different scientists, you take on different roles. You may be the person who focuses on a methodology to answer a colleague’s query, or the one who supplies the biological question. But I must say I quite enjoy developing tools that will then become available to everyone.
Such collaborations must not always be easy. For example, you are now working with biologists who do not speak the same language as you.
F. J.: It takes time on both sides, but it is very rewarding. Sometimes, it is a lack of understanding that gives rise to interesting projects. And in some cases, when you don’t understand one another, you may suddenly think of something you had not previously envisaged. A hybrid solution may emerge.
What is the next step in your work?
F. J.: Burak Yelmen has just joined our laboratory, and with the INRIA’s Tau team we have plenty of ideas. For example, we hope to find an elegant method to move from portions of genomes with several millions of base pairs to a complete artificial genome that may contain around three billion. We also wish to develop indicators to evaluate the degree of realism of artificial genomes and ensure that there is no leakage of information from real genetic material. Another part of my work with neural networks concerns their use in research on bacteria, in collaboration with Jean Cury. The aim is to rely on these statistical learning methods to follow the evolution of bacterial populations and determine the impact of events such as the introduction of new antibiotics.
Reference
"Creating artificial human genomes using generative neural networks", B. Yelmen, A. Decelle, L. Ongaro et al., 2021, PLoS Genet 17(2): e1009303. https://doi.org/10.1371/journal.pgen.1009303
- 1. CNRS researcher at the Laboratory for Computer Science (LRI) / LISN (CNRS / Université Paris-Saclay).