You are here
Life Has its Dark Matter

Might biologists have missed 85-99% of Earth’s living microorganisms? Advances in metagenomics have allowed them to show the existence of biological “dark matter.” This name is in reference to the mysterious dark matter whose mass prevents galaxies and galactic clusters from dislocation. Our biological world continues to reveal many secrets.
The three domains of life
The first classifications of life, established in the 18th century by Carl von Linné, were mainly based on morphological criteria. This system, which is efficient for large organisms such as mammals or trees, is much less efficient for classifying the entirety of life, which includes microorganisms.
So researchers turned toward universal molecular criteria. Regardless of species, all biological cells contain ribosomes, organelles that are used to synthesize proteins. Variations in the ribosomal sequences “present in all living organisms,” are used to assess distance between species and establish the large divisions of the tree of life. In the 1970s, Carl Woese established the distinction between the domains of bacteria and archaea, particularly by analyzing these molecules in prokaryotes.
Since then Eukaryotes, Bacteria, and Archaea have been considered as the three domains of life. Eukaryotic cells are characterized by the presence of a nucleus and, generally, of mitochondria. Almost all multicellular organisms fall within the eukaryote domain, which also comprises unicellular species. Bacteria and Archaea have no nucleus, but can be distinguished by the composition of their membrane.
A culture problem
To classify the multitude of microbes that they continue to discover, researchers therefore start by attempting to sequence their individual genomes. But this implies that these cell lines can be isolated and grown, which is rarely the case. Éric Bapteste, Director of Research at the IBPS,1 reminds us in his latest study2 that at least 85% of known microbes cannot be cultivated.
“The fact that microbes do not live alone makes pure culture difficult,” he explains. “They form aggregates and exist in diverse forms of symbioses. Sometimes species succeed each other over time. First, we must cultivate their precursors and reproduce the entire succession. Researchers are faced with a significant biological, biochemical, and even temporal challenge."
Various tools allow us to sequence a large part of the DNA present in a given environment, without necessarily attributing it to particular species. This is called metagenomics. These studies may apply just as well to our intestinal flora as they do to our planet's extreme nooks and crannies, such as geysers.
Researchers inventory the genetic sequences obtained in these metagenomic analyses, then subtract those that correspond to known, cultured organisms. To explore the diversity of the genetic sequences, Bapteste with colleague Philippe Lopez and their team used a two-step method. They searched in the environment for sequences that resemble those that are already known, then ran a second analysis to establish extra links.
“If we take the example of searching for relationships using family photos,” says Bapteste, “the resemblance between an individual and his grandfather will not necessarily be obvious. However links can be found if we put the child next to his father, and the father next to the grand-father. The indirect link between the individuals in these generations then becomes clear. "
Have we discovered a fourth domain?
A portion of the very numerous sequences that remain unattributed could come from new species that cannot be cultivated. Some sequences are particularly intriguing. Although they are part of known gene families, they diverge so much from sequences from cultured organisms that researchers wonder if they don’t come from organisms that are themselves very divergent, that might belong to a fourth unknown domain of life.
“Systematicians are the ones who tell us whether and when a life form becomes a new case,” Bapteste says. “Researchers use metabolic, structural, or genetic criteria but there is no fixed recipe for confirming the existence of a new domain. The process has no automatic steps; it requires arguments and consensus among the researchers. At this stage, we have no reason to say that a new domain has been discovered."
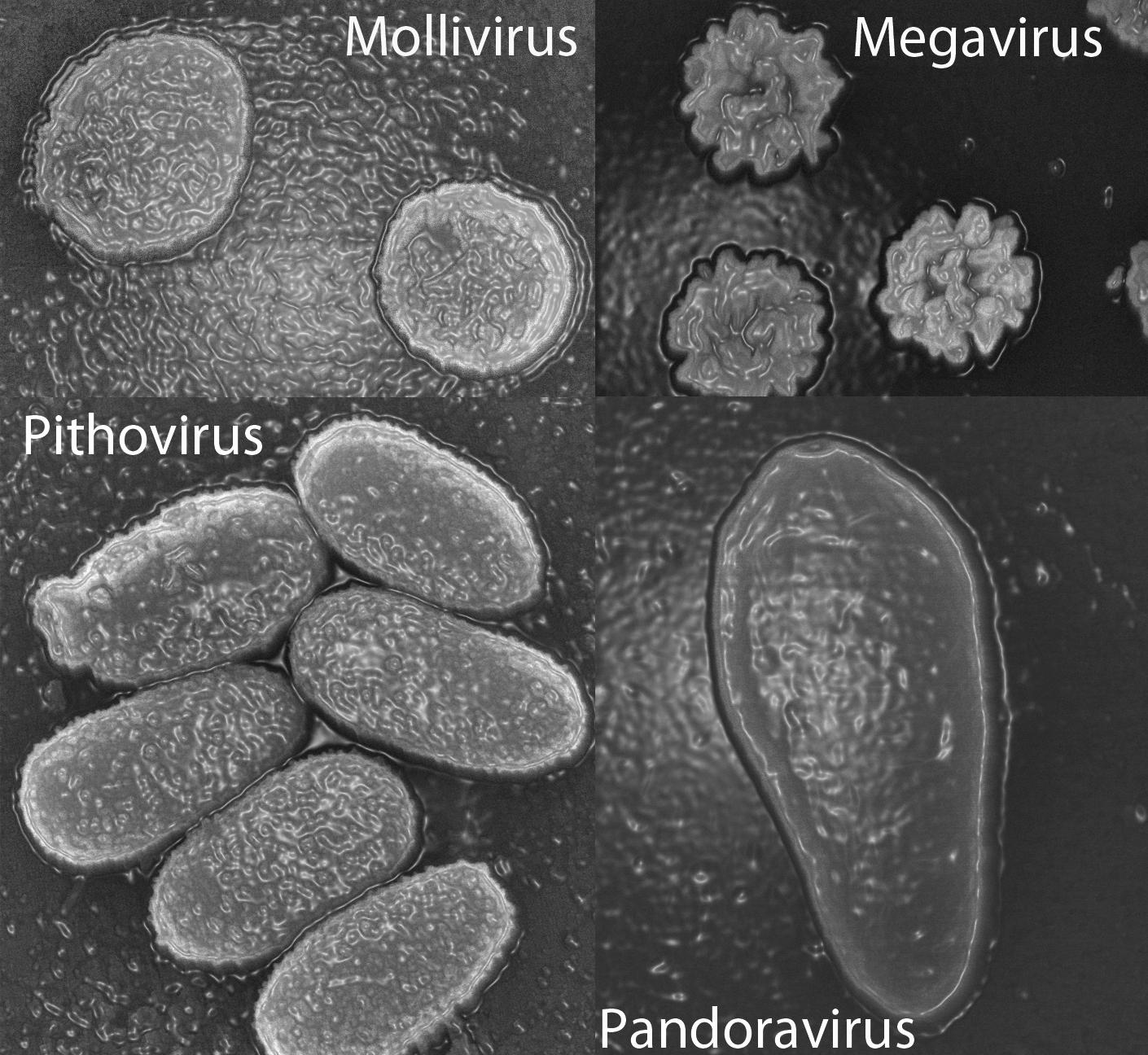
The same type of investigation followed the 2013 discovery of two giant viruses, Pandoravirus salinus and Pandoravirus dulcis. Only 7% of the genes in these viruses corresponded to known sequences, and those had no capsid (a capsid is the shell that normally surrounds a virus’ genetic material).
Purificación Lopez-Garcia, Research Director at the ESE,3 feels that phylogenetic analyses do not support the giant viruses belonging to a fourth domain of life.
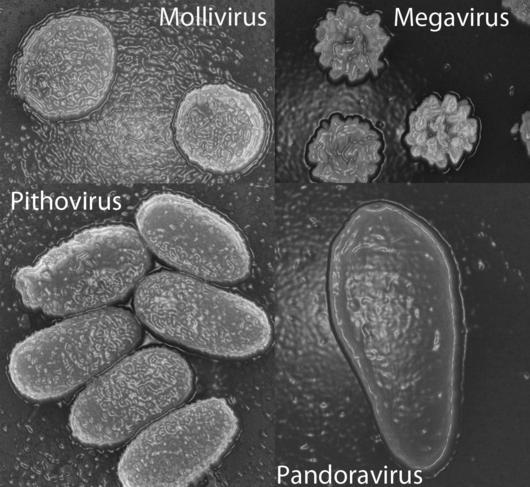
“It is tempting to state that some genes form a fourth domain," she warns, "but some can evolve very quickly, for example because of gene duplication after horizontal transfer or capture by a viral genome. They are therefore in a strange location in a phylogenetic tree but this does not imply that the organism to which they belong constitutes a completely distinct species. We cannot state anything while the genomes have not been connected to specific organisms, since for now all that has been linked to a cultured microbe or to an organism reconstructed from metagenomes does fall within the three domains of life. "
Dark matter with visible effects
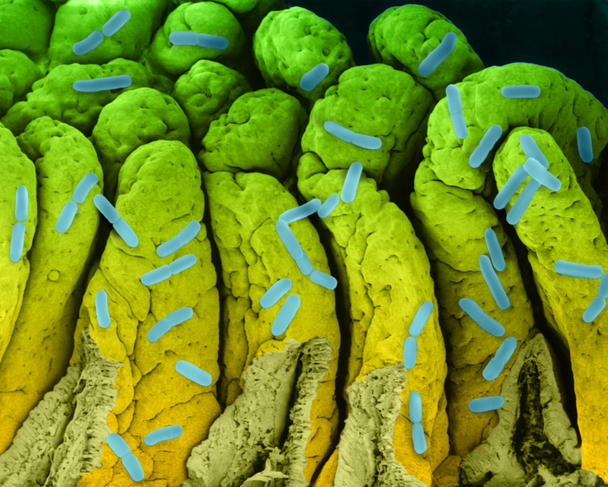
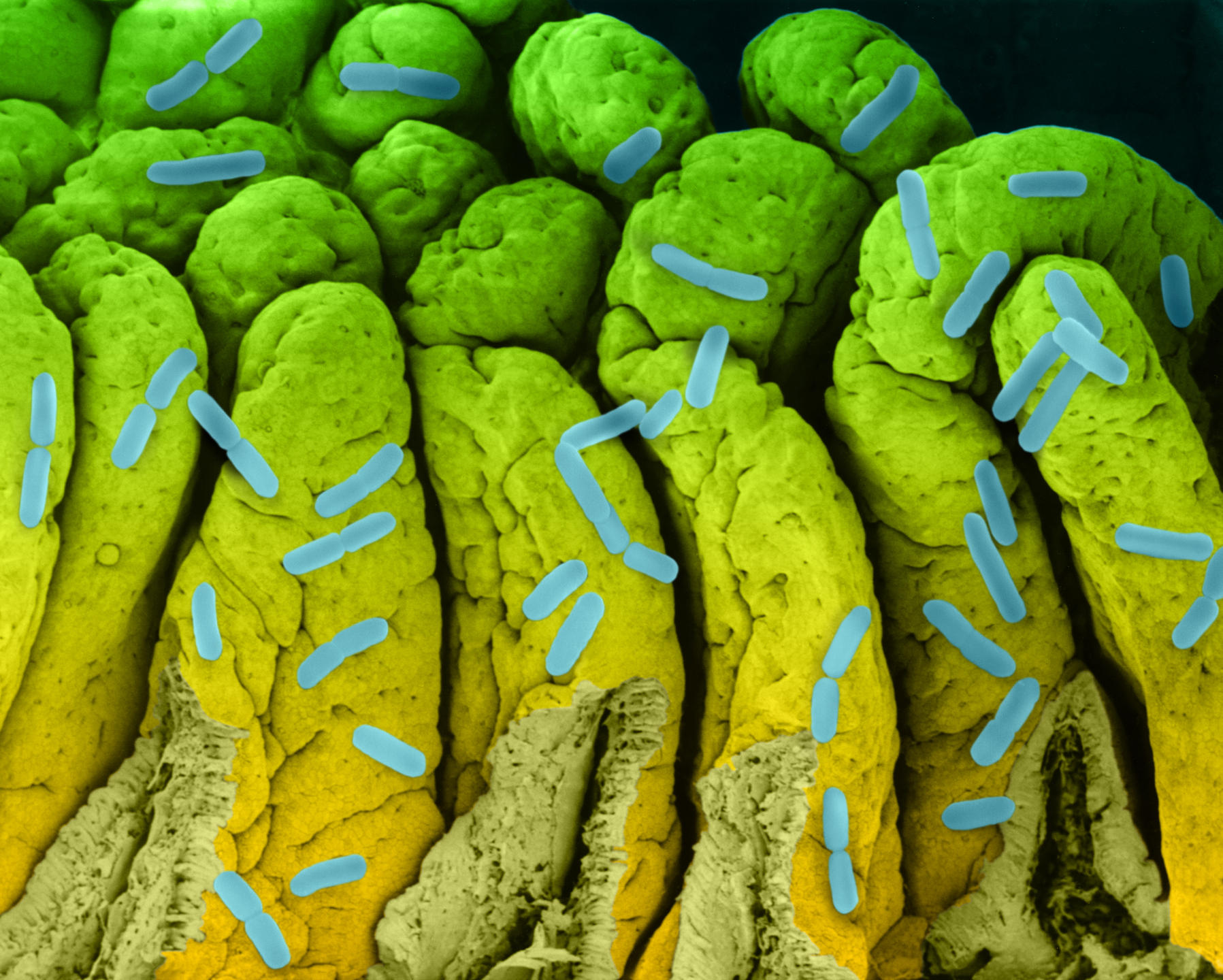
It might not have led to the inauguration of a fourth domain of life, but studying biological dark matter has proved very insightful. Dusko Ehrlich, Emeritus Research Director at the INRA MetaGénoPoliS project and Director of the Centre for Host-Microbiome Interactions at King’s College, London, has worked on the diversity of our intestinal flora. He underlines the enormous progress made since 2008 as part of the European project MetaHIT, which focuses on metagenomics.
A first study published in 2010,4 on 124 European individuals, inventoried 3.3 million intestinal microbial genes, i.e., 150 times more than the 23,000 genes in our genome. When the team increased its cohort to 1200 people, 9.9 million genes were identified. From this genetic diversity, 750 groups with more than 700 bacterial genes have been identified; 85% of the bacterial species are new.
This work has allowed us to characterize the human intestinal microbiome. Next we were able to compare the microbiomes of healthy and ill people. So a link was discovered between the composition of some microbiomes and the risk of cirrhosis.5 This has led to a 95% reliable diagnostic method based on sequencing stools. This advance could replace the very invasive liver biopsy.
Another study6 showed that the loss of diversity in the intestinal microbiome is associated with a predisposition to diabetes, hepatitis, to some cancers and to heart disease. Seven of these diseases are among the ten most common causes of death worldwide.
Ehrlich laments that “the intestinal microbiome functions as a neglected organ. In each of us, these bacteria represent a mass that may be greater than that of the brain. The public does not realize how vulnerable they are, yet they would judge this type of issue in other organs as absolutely catastrophic."
Whether it is hidden in soil, in oceans, or in our intestines, the biological role played by this so-called dark matter seems to be enormous from both the quantitative and qualitative points of view: its total mass seems to be colossal, as does its contribution to equilibrium in ecosystems, regardless of the scale being considered. This new understanding made possible by metagenomics promises to unveil biological diversity with ramifications that we cannot yet imagine.
- 1. Institut de Biologie Paris Seine. Évolution Paris Seine (CNRS / UPMC / Inserm.).
- 2. P. Lopez and al., “Highly divergent ancient gene families in metagenomic samples are compatible with additional divisions of life,” Biology Direct, 2015. 10: 64.
- 3. Laboratoire Écologie Systématique Évolution (CNRS / Université Paris Sud / AgroParisTech).
- 4. J. Qin et al., “A human gut microbial gene catalogue established by metagenomic sequencing,” Nature, 2010. 464(7285): 59-65.
- 5. J. Qin et al., “Alterations of the human gut microbiome in liver cirrhosis,” Nature, 2014. 513(7516): 59-64.
- 6. E. Le Chatelier et al., “Richness of human gut microbiome correlates with metabolic markers,” Nature,2013. 500(7464): 541–546.
Explore more
Author
A graduate from the School of Journalism in Lille, Martin Koppe has worked for a number of publications including Dossiers d’archéologie, Science et Vie Junior and La Recherche, as well the website Maxisciences.com. He also holds degrees in art history, archaeometry, and epistemology.