You are here
Mapping the Subway's Microbiome
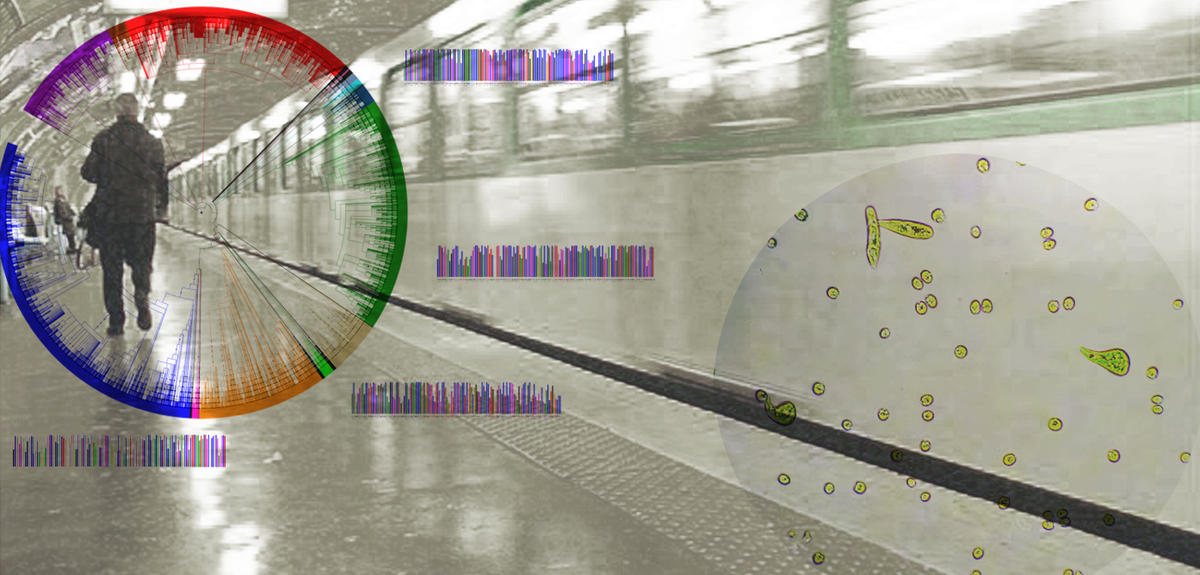
Wearing latex gloves, a man applies a large cotton swab to the steps of a central Paris metro station before sealing it in a plastic tube. Although similar to a scene from the television show CSI, no one is tracking down a dangerous criminal. The aim of the operation, repeated several times throughout the day of June 21, 2016, at the entrances of several Paris metro stations, was strictly scientific: to identify the microorganisms that have taken up residence in this suburban environment.
This unprecedented study is part of an international consortium framework dubbed MetaSUB.1 The project, which will be used to create microbial maps of the underground transit networks of the world’s largest cities, already includes 53 international megalopolises including New York, London, São Paulo, Tokyo, Berlin and Shanghai. The study in the French capital will be supervised by Hugues Richard and Ingrid Lafontaine, of the LCQB2 at the IBPS.3 “Using high throughput sequencing of the DNA samples collected on the steps of the metro, we will learn a bit more about the microbial diversity in this relatively unexplored environment,” says Alessandra Carbone, computer science professor of at the UPMC and director of LCQB.
Largely unknown bacteria
To successfully complete this ambitious investigative project, the consortium’s scientific teams are using metagenomics. At the intersection of genetics, ecology and data processing, this powerful genetic analytical method makes it possible to draw up a practically exhaustive list of the microbial species living in a given environment. “By comparing all the bacterial DNA fragments from one environment with those of species that are already categorized, metagenomics not only reveals the metabolic functions of a given community of micro-organisms, but also enables partial genome reconstruction of hitherto unknown bacterial species,” explains Carbone.
Judging by the results of the pilot campaign conducted in the New York underground between 2013 and 2015, the outcome appears very promising for the dozen other cities included in the project. Following an 18-month campaign spent sampling 468 stations and innumerable trains that crisscross one of the world’s largest metropolitan transportation networks, scientists at Weill Cornell Medical College were able to identify 562 bacterial species among the 15,000 DNA samples collected. Another highly surprising finding was that half of the samples did not belong to any species of microorganism classified to date.
Evolution underground
Although decoding this plethora of unknown genetic sequences may be extremely laborious, members of the MetaSUB project will nevertheless save enormous amounts of time using the approach developed by Carbone’s team. This highly precise analytical method based on the annotation of protein domains allows faster characterization of the functional activities of microbial communities. “To better discriminate between the different gene sequences of entirely new bacteria, our process uses information contained in the DNA sequences of known organisms throughout the entire tree of life,” says the researcher.
In each metropolitan underground transport network, MetaSUB also aims to better determine the organization and functioning of this impenetrable microcosm made up of bacterial populations. To this end, the consortium will be taking samples at regular intervals over five years in the subway stations and trains of the various cities involved. By doing this, the researchers hope to understand the effects of seasons and of certain climatic events on microbial community structures within underground transport networks. “The year when a subway station was built, its depth and its humidity levels are all factors likely to affect this bacterial diversity,” stresses Carbone. “For example, in one New York underground station flooded in November 2012 during Hurricane Sandy, our US colleagues were able to identify 10 species of bacteria that normally dwell in marine environments.”
New therapeutic strategies on the horizon
In Paris, the LCQB is waiting for the regional transport authority, RATP, to greenlight initial sampling operations within its network, rather than just at station entrances. Once authorizations have been granted for each of the 16 Paris metro lines, tens of computer and biology students at the UPMC involved in the MetaSUB project will be able to collect bacterial samples from both platforms and trains within the metro. “By involving our students in a participatory science project like this one, we are preparing them for the type of biology and ecology problems set to emerge in the coming decades,” enthuses Carbone. Airborne bacteria will also be captured by means of filters in the ventilation systems.
By crosschecking all of these data with specific characteristics for each station (cleaning dates, daily passenger traffic, air quality data, etc.), scientists hope to be establish links between the composition of underground bacterial populations and the presence of atmospheric pollutants and allergenic substances. In particular, such analyses could help improve our understanding of how pollution impacts the health of the city’s inhabitants. Ultimately, this unprecedented collection of information should enable the identification of new biosynthetic pathways in bacteria. This could provide an opportunity to discover new medicines or new antibiotic-resistant genes.
- 1. The MetaSUB International Consortium, “The Metagenomics and Metadesign of the Subways and Urban Biomes (MetaSUB) International Consortium inaugural meeting report”, Microbiome, 2016. 4: 24.
- 2. laboratoire Biologie computationnelle et quantitative (CNRS / UPMC).
- 3. Institut de Biologie Paris-Seine (CNRS / UPMC / Inserm).
Explore more
Author
After first studying biology, Grégory Fléchet graduated with a master of science journalism. His areas of interest include ecology, the environment and health. From Saint-Etienne, he moved to Paris in 2007, where he now works as a freelance journalist.










